Molecular mechanisms of RNA-triggered gene silencing machineries
- PMID: 22304792
- PMCID: PMC3398549
- DOI: 10.1021/ar200253u
Molecular mechanisms of RNA-triggered gene silencing machineries
Abstract
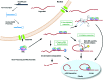
Gene silencing by RNA triggers is an ancient, evolutionarily conserved, and widespread phenomenon. This process, known as RNA interference (RNAi), occurs when double-stranded RNA helices induce cleavage of their complementary mRNAs. Because these RNA molecules can be introduced exogenously as small interfering RNAs (siRNAs), RNAi has become an everyday experimental tool in laboratory research. In addition, the number of RNA-based therapeutics that are currently in clinical trials for a variety of human diseases demonstrate the therapeutic potential of RNAi. In this Account, we focus on our current understanding of the structure and function of various classes of RNAi triggers and how this knowledge has contributed to our understanding of the biogenesis and catalytic functions of siRNA and microRNA in mammalian cells. Mechanistic studies to understand the structure and function of small RNAs that induce RNAi have illuminated broad functions of the ancient RNAi machinery in animals and plants. In addition, such studies have provided insight to identify endogenous physiological gene silencing RNA triggers that engage functional machineries similar to siRNAs. Several endogenous small RNA species have been identified: small noncoding RNAs (microRNAs), piwi-interacting RNAs (piRNAs), and endogenous siRNAs (endo-siRNAs). microRNAs are the most widespread class of small RNAs in mammalian cells. Despite their importance in biology and medicine, the molecular and cellular mechanisms of microRNA biogenesis and function are not fully understood. We provide an overview of the current understanding of how these molecules are synthesized within cells and how they act on gene targets. Interesting questions remain both for understanding the effects of modifications and editing on microRNAs and the interactions between microRNAs and other cellular RNAs such as long noncoding RNAs.
Figures





References
-
- Fire A.; Xu S.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. - PubMed
-
- Kim V. N.; Han J.; Siomi M. C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. - PubMed
-
- Krol J.; Loedige I.; Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. - PubMed
-
- Bagga S.; Bracht J.; Hunter S.; Massirer K.; Holtz J.; Eachus R.; Pasquinelli A. E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005, 122, 553–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

