Analysis of the SOS response of Vibrio and other bacteria with multiple chromosomes
- PMID: 22305460
- PMCID: PMC3323433
- DOI: 10.1186/1471-2164-13-58
Analysis of the SOS response of Vibrio and other bacteria with multiple chromosomes
Abstract
Background: The SOS response is a well-known regulatory network present in most bacteria and aimed at addressing DNA damage. It has also been linked extensively to stress-induced mutagenesis, virulence and the emergence and dissemination of antibiotic resistance determinants. Recently, the SOS response has been shown to regulate the activity of integrases in the chromosomal superintegrons of the Vibrionaceae, which encompasses a wide range of pathogenic species harboring multiple chromosomes. Here we combine in silico and in vitro techniques to perform a comparative genomics analysis of the SOS regulon in the Vibrionaceae, and we extend the methodology to map this transcriptional network in other bacterial species harboring multiple chromosomes.
Results: Our analysis provides the first comprehensive description of the SOS response in a family (Vibrionaceae) that includes major human pathogens. It also identifies several previously unreported members of the SOS transcriptional network, including two proteins of unknown function. The analysis of the SOS response in other bacterial species with multiple chromosomes uncovers additional regulon members and reveals that there is a conserved core of SOS genes, and that specialized additions to this basic network take place in different phylogenetic groups. Our results also indicate that across all groups the main elements of the SOS response are always found in the large chromosome, whereas specialized additions are found in the smaller chromosomes and plasmids.
Conclusions: Our findings confirm that the SOS response of the Vibrionaceae is strongly linked with pathogenicity and dissemination of antibiotic resistance, and suggest that the characterization of the newly identified members of this regulon could provide key insights into the pathogenesis of Vibrio. The persistent location of key SOS genes in the large chromosome across several bacterial groups confirms that the SOS response plays an essential role in these organisms and sheds light into the mechanisms of evolution of global transcriptional networks involved in adaptability and rapid response to environmental changes, suggesting that small chromosomes may act as evolutionary test beds for the rewiring of transcriptional networks.
Figures

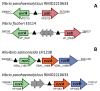


Similar articles
-
Characterization of the SOS meta-regulon in the human gut microbiome.Bioinformatics. 2014 May 1;30(9):1193-7. doi: 10.1093/bioinformatics/btt753. Epub 2014 Jan 8. Bioinformatics. 2014. PMID: 24407225 Free PMC article.
-
An SOS Regulon under Control of a Noncanonical LexA-Binding Motif in the Betaproteobacteria.J Bacteriol. 2015 Aug;197(16):2622-30. doi: 10.1128/JB.00035-15. Epub 2015 May 18. J Bacteriol. 2015. PMID: 25986903 Free PMC article.
-
Computational analysis of LexA regulons in Cyanobacteria.BMC Genomics. 2010 Sep 29;11:527. doi: 10.1186/1471-2164-11-527. BMC Genomics. 2010. PMID: 20920248 Free PMC article.
-
Aeons of distress: an evolutionary perspective on the bacterial SOS response.FEMS Microbiol Rev. 2007 Nov;31(6):637-56. doi: 10.1111/j.1574-6976.2007.00082.x. Epub 2007 Sep 18. FEMS Microbiol Rev. 2007. PMID: 17883408 Review.
-
Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon.Mol Microbiol. 2006 Dec;62(5):1228-38. doi: 10.1111/j.1365-2958.2006.05444.x. Epub 2006 Oct 17. Mol Microbiol. 2006. PMID: 17042786 Review.
Cited by
-
Elevated mutagenesis does not explain the increased frequency of antibiotic resistant mutants in starved aging colonies.PLoS Genet. 2013 Nov;9(11):e1003968. doi: 10.1371/journal.pgen.1003968. Epub 2013 Nov 14. PLoS Genet. 2013. PMID: 24244205 Free PMC article.
-
On or Off: Life-Changing Decisions Made by Vibrio cholerae Under Stress.Infect Microbes Dis. 2020 Oct 14;2(4):127-135. doi: 10.1097/IM9.0000000000000037. eCollection 2020 Dec. Infect Microbes Dis. 2020. PMID: 38630076 Free PMC article. Review.
-
Characterization of the SOS meta-regulon in the human gut microbiome.Bioinformatics. 2014 May 1;30(9):1193-7. doi: 10.1093/bioinformatics/btt753. Epub 2014 Jan 8. Bioinformatics. 2014. PMID: 24407225 Free PMC article.
-
Identification of EppR, a Second Repressor of Error-Prone DNA Polymerase Genes in Acinetobacter baumannii.Mol Microbiol. 2025 Jul;124(1):20-39. doi: 10.1111/mmi.15368. Epub 2025 Apr 19. Mol Microbiol. 2025. PMID: 40251897 Free PMC article.
-
Potential mechanisms of attenuation for rifampicin-passaged strains of Flavobacterium psychrophilum.BMC Microbiol. 2015 Sep 16;15:179. doi: 10.1186/s12866-015-0518-1. BMC Microbiol. 2015. PMID: 26377311 Free PMC article.
References
-
- Radman M. In: Molecular and environmental aspects of mutagenesis. Prokash L, Sherman F, Miller M, Lawrence C, Tabor HW: Charles C, editor. Thomas Publisher, Springfield, IL; 1974. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis; pp. 128–142.
-
- Walker GC. In: Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Neidhart FC, Ingram JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editor. Vol. 1. Washington, D.C.: American Society for Microbiology; 1987. The SOS response of Escherichia coli.
-
- Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Molecular microbiology. 2000;35:1560–1572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

