Small-molecule-induced DNA damage identifies alternative DNA structures in human genes
- PMID: 22306580
- PMCID: PMC3433707
- DOI: 10.1038/nchembio.780
Small-molecule-induced DNA damage identifies alternative DNA structures in human genes
Abstract
Guanine-rich DNA sequences that can adopt non-Watson-Crick structures in vitro are prevalent in the human genome. Whether such structures normally exist in mammalian cells has, however, been the subject of active research for decades. Here we show that the G-quadruplex-interacting drug pyridostatin promotes growth arrest in human cancer cells by inducing replication- and transcription-dependent DNA damage. A chromatin immunoprecipitation sequencing analysis of the DNA damage marker γH2AX provided the genome-wide distribution of pyridostatin-induced sites of damage and revealed that pyridostatin targets gene bodies containing clusters of sequences with a propensity for G-quadruplex formation. As a result, pyridostatin modulated the expression of these genes, including the proto-oncogene SRC. We observed that pyridostatin reduced SRC protein abundance and SRC-dependent cellular motility in human breast cancer cells, validating SRC as a target of this drug. Our unbiased approach to define genomic sites of action for a drug establishes a framework for discovering functional DNA-drug interactions.
Figures
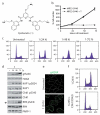
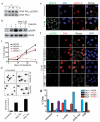

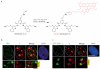
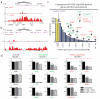

Comment in
-
Alternative DNA structures: G4 DNA in cells: itae missa est?Nat Chem Biol. 2012 Feb 15;8(3):225-6. doi: 10.1038/nchembio.793. Nat Chem Biol. 2012. PMID: 22337092 No abstract available.
References
-
- Dervan PB. Design of sequence-specific DNA-binding molecules. Science. 1986;232:464–71. - PubMed
-
- Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–224. - PubMed
-
- Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387:202–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

