DNase I sensitivity QTLs are a major determinant of human expression variation
- PMID: 22307276
- PMCID: PMC3501342
- DOI: 10.1038/nature10808
DNase I sensitivity QTLs are a major determinant of human expression variation
Abstract
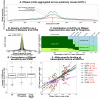
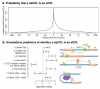
The mapping of expression quantitative trait loci (eQTLs) has emerged as an important tool for linking genetic variation to changes in gene regulation. However, it remains difficult to identify the causal variants underlying eQTLs, and little is known about the regulatory mechanisms by which they act. Here we show that genetic variants that modify chromatin accessibility and transcription factor binding are a major mechanism through which genetic variation leads to gene expression differences among humans. We used DNase I sequencing to measure chromatin accessibility in 70 Yoruba lymphoblastoid cell lines, for which genome-wide genotypes and estimates of gene expression levels are also available. We obtained a total of 2.7 billion uniquely mapped DNase I-sequencing (DNase-seq) reads, which allowed us to produce genome-wide maps of chromatin accessibility for each individual. We identified 8,902 locations at which the DNase-seq read depth correlated significantly with genotype at a nearby single nucleotide polymorphism or insertion/deletion (false discovery rate = 10%). We call such variants 'DNase I sensitivity quantitative trait loci' (dsQTLs). We found that dsQTLs are strongly enriched within inferred transcription factor binding sites and are frequently associated with allele-specific changes in transcription factor binding. A substantial fraction (16%) of dsQTLs are also associated with variation in the expression levels of nearby genes (that is, these loci are also classified as eQTLs). Conversely, we estimate that as many as 55% of eQTL single nucleotide polymorphisms are also dsQTLs. Our observations indicate that dsQTLs are highly abundant in the human genome and are likely to be important contributors to phenotypic variation.
Figures


References
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–5. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

