Equine stomachs harbor an abundant and diverse mucosal microbiota
- PMID: 22307294
- PMCID: PMC3318809
- DOI: 10.1128/AEM.06252-11
Equine stomachs harbor an abundant and diverse mucosal microbiota
Abstract
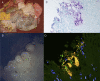
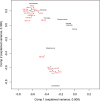
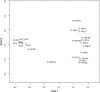
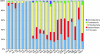
Little is known about the gastric mucosal microbiota in healthy horses, and its role in gastric disease has not been critically examined. The present study used a combination of 16S rRNA bacterial tag-encoded pyrosequencing (bTEFAP) and fluorescence in situ hybridization (FISH) to characterize the composition and spatial distribution of selected gastric mucosal microbiota of healthy horses. Biopsy specimens of the squamous, glandular, antral, and any ulcerated mucosa were obtained from 6 healthy horses by gastroscopy and from 3 horses immediately postmortem. Pyrosequencing was performed on biopsy specimens from 6 of the horses and yielded 53,920 reads in total, with 631 to 4,345 reads in each region per horse. The microbiome segregated into two distinct clusters comprised of horses that were stabled, fed hay, and sampled at postmortem (cluster 1) and horses that were pastured on grass, fed hay, and biopsied gastroscopically after a 12-h fast (cluster 2). The types of bacteria obtained from different anatomic regions clustered by horse rather than region. The dominant bacteria in cluster 1 were Firmicutes (>83% reads/sample), mainly Streptococcus spp., Lactobacillus spp. and, Sarcina spp. Cluster 2 was more diverse, with predominantly Proteobacteria, Bacteroidetes, and Firmicutes, consisting of Actinobacillus spp. Moraxella spp., Prevotella spp., and Porphyromonas spp. Helicobacter sp. sequences were not identified in any of 53,920 reads. FISH (n = 9) revealed bacteria throughout the stomach in close apposition to the mucosa, with significantly more Streptococcus spp. present in the glandular region of the stomach. The equine stomach harbors an abundant and diverse mucosal microbiota that varies by individual.
Figures


References
-
- Acosta-Martinez V, Dowd SE, Sun Y, Allen V. 2009. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 4:2762–2770
-
- Alexander F. 1972. Certain aspects of the physiology and pharmacology of the horse's digestive tract. Equine Vet. J. 4:166–169
-
- Al Jassim RAM, Scott T, Trebbin AL, Trott D, Pollitt CC. 2005. The genetic diversity of lactic acid producing bacteria in the equine gastrointestinal tract. FEMS Microbiol. Lett. 248:75–81 - PubMed
-
- Al Jassim RAM. 2006. Supplementary feeding of horses with processed sorghum grains and oats. Anim. Feed Sci. Tech. 125:33–44
-
- Andrews FM, Buchanan BR, Elliot SB, Clairday NA, Edwards LH. 2005. Gastric ulcers in horses. J. Anim. Sci. 83:E18–E21
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

