NAD(P)+-malic enzyme mutants of Sinorhizobium sp. strain NGR234, but not Azorhizobium caulinodans ORS571, maintain symbiotic N2 fixation capabilities
- PMID: 22307295
- PMCID: PMC3318798
- DOI: 10.1128/AEM.06412-11
NAD(P)+-malic enzyme mutants of Sinorhizobium sp. strain NGR234, but not Azorhizobium caulinodans ORS571, maintain symbiotic N2 fixation capabilities
Abstract
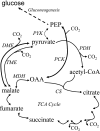
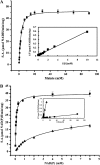
C(4)-dicarboxylic acids appear to be metabolized via the tricarboxylic acid (TCA) cycle in N(2)-fixing bacteria (bacteroids) within legume nodules. In Sinorhizobium meliloti bacteroids from alfalfa, NAD(+)-malic enzyme (DME) is required for N(2) fixation, and this activity is thought to be required for the anaplerotic synthesis of pyruvate. In contrast, in the pea symbiont Rhizobium leguminosarum, pyruvate synthesis occurs via either DME or a pathway catalyzed by phosphoenolpyruvate carboxykinase (PCK) and pyruvate kinase (PYK). Here we report that dme mutants of the broad-host-range Sinorhizobium sp. strain NGR234 formed nodules whose level of N(2) fixation varied from 27 to 83% (plant dry weight) of the wild-type level, depending on the host plant inoculated. NGR234 bacteroids had significant PCK activity, and while single pckA and single dme mutants fixed N(2) at reduced rates, a pckA dme double mutant had no N(2)-fixing activity (Fix(-)). Thus, NGR234 bacteroids appear to synthesize pyruvate from TCA cycle intermediates via DME or PCK pathways. These NGR234 data, together with other reports, suggested that the completely Fix(-) phenotype of S. meliloti dme mutants may be specific to the alfalfa-S. meliloti symbiosis. We therefore examined the ME-like genes azc3656 and azc0119 from Azorhizobium caulinodans, as azc3656 mutants were previously shown to form Fix(-) nodules on the tropical legume Sesbania rostrata. We found that purified AZC3656 protein is an NAD(P)(+)-malic enzyme whose activity is inhibited by acetyl-coenzyme A (acetyl-CoA) and stimulated by succinate and fumarate. Thus, whereas DME is required for symbiotic N(2) fixation in A. caulinodans and S. meliloti, in other rhizobia this activity can be bypassed via another pathway(s).
Figures


Similar articles
-
A Chemotaxis Receptor Modulates Nodulation during the Azorhizobium caulinodans-Sesbania rostrata Symbiosis.Appl Environ Microbiol. 2016 May 16;82(11):3174-84. doi: 10.1128/AEM.00230-16. Print 2016 Jun 1. Appl Environ Microbiol. 2016. PMID: 26994081 Free PMC article.
-
Properties of NAD(+)- and NADP(+)-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti and differential expression of their genes in nitrogen-fixing bacteroids.Microbiology (Reading). 1997 Feb;143 ( Pt 2):489-498. doi: 10.1099/00221287-143-2-489. Microbiology (Reading). 1997. PMID: 9043124
-
Pyruvate is synthesized by two pathways in pea bacteroids with different efficiencies for nitrogen fixation.J Bacteriol. 2010 Oct;192(19):4944-53. doi: 10.1128/JB.00294-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675477 Free PMC article.
-
Malic enzyme cofactor and domain requirements for symbiotic N2 fixation by Sinorhizobium meliloti.J Bacteriol. 2007 Jan;189(1):160-8. doi: 10.1128/JB.01425-06. Epub 2006 Oct 27. J Bacteriol. 2007. PMID: 17071765 Free PMC article.
-
Tricarboxylic acid cycle and anaplerotic enzymes in rhizobia.FEMS Microbiol Rev. 1998 Jun;22(2):105-23. doi: 10.1111/j.1574-6976.1998.tb00363.x. FEMS Microbiol Rev. 1998. PMID: 9729766 Review.
Cited by
-
Role of O2 in the Growth of Rhizobium leguminosarum bv. viciae 3841 on Glucose and Succinate.J Bacteriol. 2016 Dec 13;199(1):e00572-16. doi: 10.1128/JB.00572-16. Print 2017 Jan 1. J Bacteriol. 2016. PMID: 27795326 Free PMC article.
-
A minimized symbiotic gene set from the 1.68 Mb pSymB chromid of Sinorhizobium meliloti reveals auxiliary symbiotic loci.BMC Biol. 2025 Jul 9;23(1):204. doi: 10.1186/s12915-025-02298-5. BMC Biol. 2025. PMID: 40629387 Free PMC article.
-
Mechanisms for Generating Low Potential Electrons across the Metabolic Diversity of Nitrogen-Fixing Bacteria.Appl Environ Microbiol. 2023 May 31;89(5):e0037823. doi: 10.1128/aem.00378-23. Epub 2023 May 8. Appl Environ Microbiol. 2023. PMID: 37154716 Free PMC article. Review.
-
Interaction and Regulation of Carbon, Nitrogen, and Phosphorus Metabolisms in Root Nodules of Legumes.Front Plant Sci. 2018 Dec 18;9:1860. doi: 10.3389/fpls.2018.01860. eCollection 2018. Front Plant Sci. 2018. PMID: 30619423 Free PMC article. Review.
-
An enzyme-centric approach for constructing an amperometric l-malate biosensor with a long and programmable linear range.Protein Sci. 2023 Sep;32(9):e4743. doi: 10.1002/pro.4743. Protein Sci. 2023. PMID: 37515423 Free PMC article.
References
-
- Arwas R, McKay IA, Rowney FRP, Dilworth MJ, Glenn AR. 1985. Properties of organic-acid utilization mutants of Rhizobium leguminosarum strain-300. J. Gen. Microbiol. 131:2059–2066
-
- Bala A, Giller KE. 2001. Symbiotic specificity of tropical tree rhizobia for host legumes. New Phytol. 149:495–507 - PubMed
-
- Blasing OE, Westhoff P, Svensson P. 2000. Evolution of C4 phospho-enolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. J. Biol. Chem. 275:27917–27923 - PubMed
-
- Reference deleted.
-
- Bradford MM. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248–254 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

