Accelerated evolution and coevolution drove the evolutionary history of AGPase sub-units during angiosperm radiation
- PMID: 22307567
- PMCID: PMC3286274
- DOI: 10.1093/aob/mcr303
Accelerated evolution and coevolution drove the evolutionary history of AGPase sub-units during angiosperm radiation
Abstract
Background and aims: ADP-glucose pyrophosphorylase (AGPase) is a key enzyme of starch biosynthesis. In the green plant lineage, it is composed of two large (LSU) and two small (SSU) sub-units encoded by paralogous genes, as a consequence of several rounds of duplication. First, our aim was to detect specific patterns of molecular evolution following duplication events and the divergence between monocotyledons and dicotyledons. Secondly, we investigated coevolution between amino acids both within and between sub-units.
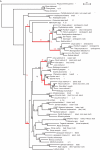
Methods: A phylogeny of each AGPase sub-unit was built using all gymnosperm and angiosperm sequences available in databases. Accelerated evolution along specific branches was tested using the ratio of the non-synonymous to the synonymous substitution rate. Coevolution between amino acids was investigated taking into account compensatory changes between co-substitutions.
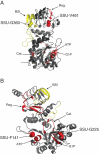
Key results: We showed that SSU paralogues evolved under high functional constraints during angiosperm radiation, with a significant level of coevolution between amino acids that participate in SSU major functions. In contrast, in the LSU paralogues, we identified residues under positive selection (1) following the first LSU duplication that gave rise to two paralogues mainly expressed in angiosperm source and sink tissues, respectively; and (2) following the emergence of grass-specific paralogues expressed in the endosperm. Finally, we found coevolution between residues that belong to the interaction domains of both sub-units.
Conclusions: Our results support the view that coevolution among amino acid residues, especially those lying in the interaction domain of each sub-unit, played an important role in AGPase evolution. First, within SSU, coevolution allowed compensating mutations in a highly constrained context. Secondly, the LSU paralogues probably acquired tissue-specific expression and regulatory properties via the coevolution between sub-unit interacting domains. Finally, the pattern we observed during LSU evolution is consistent with repeated sub-functionalization under 'Escape from Adaptive Conflict', a model rarely illustrated in the literature.
Figures


Similar articles
-
Molecular evolution accompanying functional divergence of duplicated genes along the plant starch biosynthesis pathway.BMC Evol Biol. 2014 May 15;14:103. doi: 10.1186/1471-2148-14-103. BMC Evol Biol. 2014. PMID: 24884572 Free PMC article.
-
Contrasted patterns of selection since maize domestication on duplicated genes encoding a starch pathway enzyme.Theor Appl Genet. 2011 Mar;122(4):705-22. doi: 10.1007/s00122-010-1480-9. Theor Appl Genet. 2011. PMID: 21060986
-
Duplications and functional divergence of ADP-glucose pyrophosphorylase genes in plants.BMC Evol Biol. 2008 Aug 12;8:232. doi: 10.1186/1471-2148-8-232. BMC Evol Biol. 2008. PMID: 18700010 Free PMC article.
-
The evolution of the starch biosynthetic pathway in cereals and other grasses.J Exp Bot. 2009;60(9):2481-92. doi: 10.1093/jxb/erp141. J Exp Bot. 2009. PMID: 19505928 Review.
-
Convergent evolution in angiosperms adapted to cold climates.Plant Commun. 2025 Feb 10;6(2):101258. doi: 10.1016/j.xplc.2025.101258. Epub 2025 Jan 23. Plant Commun. 2025. PMID: 39849842 Free PMC article. Review.
Cited by
-
AGPase: its role in crop productivity with emphasis on heat tolerance in cereals.Theor Appl Genet. 2015 Oct;128(10):1893-916. doi: 10.1007/s00122-015-2565-2. Epub 2015 Jul 8. Theor Appl Genet. 2015. PMID: 26152573 Review.
-
Decreased Nitrogen and Carbohydrate Metabolism Activity Leads to Grain Yield Reduction in Qingke Under Continuous Cropping.Plants (Basel). 2025 Jul 19;14(14):2235. doi: 10.3390/plants14142235. Plants (Basel). 2025. PMID: 40733472 Free PMC article.
-
Molecular evolution accompanying functional divergence of duplicated genes along the plant starch biosynthesis pathway.BMC Evol Biol. 2014 May 15;14:103. doi: 10.1186/1471-2148-14-103. BMC Evol Biol. 2014. PMID: 24884572 Free PMC article.
-
Starch Synthesis-Related Genes (SSRG) Evolution in the Genus Oryza.Plants (Basel). 2021 May 25;10(6):1057. doi: 10.3390/plants10061057. Plants (Basel). 2021. PMID: 34070565 Free PMC article.
-
Comparative Analysis of AGPase Genes and Encoded Proteins in Eight Monocots and Three Dicots with Emphasis on Wheat.Front Plant Sci. 2017 Jan 24;8:19. doi: 10.3389/fpls.2017.00019. eCollection 2017. Front Plant Sci. 2017. PMID: 28174576 Free PMC article.
References
-
- Aguileta G, Bielawski JP, Yang Z. Evolutionary rate variation among vertebrate beta globin genes: implications for dating gene family duplication events. Gene. 2006;380:21–29. - PubMed
-
- Altschuh D, Lesk AM, Bloomer AC, Klug A. Correlation of co-ordinated amino acid substitutions with function in viruses related to tobacco mosaic virus. Journal of Molecular Biology. 1987;193:693–707. - PubMed
-
- Atchley WR, Wollenberg KR, Fitch WM, Terhalle W, Dress AW. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Molecular Biology and Evolution. 2000;17:164–178. - PubMed

