Transcriptional interpretation of the EGF receptor signaling gradient
- PMID: 22307613
- PMCID: PMC3277169
- DOI: 10.1073/pnas.1115190109
Transcriptional interpretation of the EGF receptor signaling gradient
Abstract
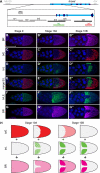
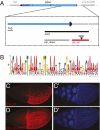
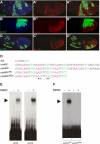
Epidermal growth factor receptor (EGFR) controls a wide range of developmental events, from body axes specification in insects to cardiac development in humans. During Drosophila oogenesis, a gradient of EGFR activation patterns the follicular epithelium. Multiple transcriptional targets of EGFR in this tissue have been identified, but their regulatory elements are essentially unknown. We report the regulatory elements of broad (br) and pipe (pip), two important targets of EGFR signaling in Drosophila oogenesis. br is expressed in a complex pattern that prefigures the formation of respiratory eggshell appendages. We found that this pattern is generated by dynamic activities of two regulatory elements, which display different responses to Pointed, Capicua, and Mirror, transcription factors involved in the EGFR-mediated gene expression. One of these elements is active in a pattern similar to pip, a gene repressed by EGFR and essential for establishing the dorsoventral polarity of the embryo. We demonstrate that this similarity of expression depends on a common sequence motif that binds Mirror in vitro and is essential for transcriptional repression in vivo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Dynamic model for the coordination of two enhancers of broad by EGFR signaling.Proc Natl Acad Sci U S A. 2013 Oct 29;110(44):17939-44. doi: 10.1073/pnas.1304753110. Epub 2013 Oct 14. Proc Natl Acad Sci U S A. 2013. PMID: 24127599 Free PMC article.
-
Feedback control of the EGFR signaling gradient: superposition of domain-splitting events in Drosophila oogenesis.Development. 2009 Sep;136(17):2903-11. doi: 10.1242/dev.039545. Epub 2009 Jul 29. Development. 2009. PMID: 19641013 Free PMC article.
-
EGFR-dependent downregulation of Capicua and the establishment of Drosophila dorsoventral polarity.Fly (Austin). 2012 Oct-Dec;6(4):234-9. doi: 10.4161/fly.21160. Epub 2012 Aug 10. Fly (Austin). 2012. PMID: 22878648 Free PMC article.
-
EGF receptor signaling in Drosophila oogenesis.Curr Top Dev Biol. 1999;44:203-43. doi: 10.1016/s0070-2153(08)60471-8. Curr Top Dev Biol. 1999. PMID: 9891881 Review.
-
Versatility in signalling: multiple responses to EGF receptor activation during Drosophila oogenesis.Trends Cell Biol. 1999 Jan;9(1):1-4. doi: 10.1016/s0962-8924(98)01413-5. Trends Cell Biol. 1999. PMID: 10087609 Review.
Cited by
-
Dynamic model for the coordination of two enhancers of broad by EGFR signaling.Proc Natl Acad Sci U S A. 2013 Oct 29;110(44):17939-44. doi: 10.1073/pnas.1304753110. Epub 2013 Oct 14. Proc Natl Acad Sci U S A. 2013. PMID: 24127599 Free PMC article.
-
A Unifying Framework for Understanding Biological Structures and Functions Across Levels of Biological Organization.Integr Comp Biol. 2022 Feb 5;61(6):2038-2047. doi: 10.1093/icb/icab167. Integr Comp Biol. 2022. PMID: 34302339 Free PMC article.
-
Shared cis-regulatory modules control expression of the tandem paralogs midline and H15 in the follicular epithelium.Development. 2022 Nov 15;149(22):dev201016. doi: 10.1242/dev.201016. Epub 2022 Nov 16. Development. 2022. PMID: 36278857 Free PMC article.
-
The Capicua repressor--a general sensor of RTK signaling in development and disease.J Cell Sci. 2012 Mar 15;125(Pt 6):1383-91. doi: 10.1242/jcs.092965. J Cell Sci. 2012. PMID: 22526417 Free PMC article. Review.
-
Stochastic phenotypes in RAS-dependent developmental diseases.Curr Biol. 2023 Mar 13;33(5):807-816.e4. doi: 10.1016/j.cub.2023.01.008. Epub 2023 Jan 26. Curr Biol. 2023. PMID: 36706752 Free PMC article.
References
-
- Moghal N, Sternberg PW. The epidermal growth factor system in Caenorhabditis elegans. Exp Cell Res. 2003;284:150–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

