The TonB3 system in the human pathogen Vibrio vulnificus is under the control of the global regulators Lrp and cyclic AMP receptor protein
- PMID: 22307757
- PMCID: PMC3318495
- DOI: 10.1128/JB.06614-11
The TonB3 system in the human pathogen Vibrio vulnificus is under the control of the global regulators Lrp and cyclic AMP receptor protein
Abstract
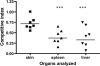
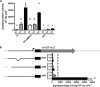
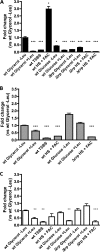
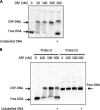
TonB systems transduce the proton motive force of the cytoplasmic membrane to energize substrate transport through a specific TonB-dependent transporter across the outer membrane. Vibrio vulnificus, an opportunistic marine pathogen that can cause a fatal septicemic disease in humans and eels, possesses three TonB systems. While the TonB1 and TonB2 systems are iron regulated, the TonB3 system is induced when the bacterium grows in human serum. In this work we have determined the essential roles of the leucine-responsive protein (Lrp) and cyclic AMP (cAMP) receptor protein (CRP) in the transcriptional activation of this system. Whereas Lrp shows at least four very distinctive DNA binding regions spread out from position -59 to -509, cAMP-CRP binds exclusively in a region centered at position -122.5 from the start point of the transcription. Our results suggest that both proteins bind simultaneously to the region closer to the RNA polymerase binding site. Importantly, we report that the TonB3 system is induced not only by serum but also during growth in minimal medium with glycerol as the sole carbon source and low concentrations of Casamino Acids. In addition to catabolite repression by glucose, l-leucine acts by inhibiting the binding of Lrp to the promoter region, hence preventing transcription of the TonB3 operon. Thus, this TonB system is under the direct control of two global regulators that can integrate different environmental signals (i.e., glucose starvation and the transition between "feast" and "famine"). These results shed light on new mechanisms of regulation for a TonB system that could be widespread in other organisms.
Figures







References
-
- Brennt CE, Wright AC, Dutta SK, Morris JGJ. 1991. Growth of Vibrio vulnificus in serum from alcoholics: association with high transferrin iron saturation. J. Infect. Dis. 164: 1030– 1032 - PubMed
-
- Browning DF, et al. 2004. Modulation of CRP-dependent transcription at the Escherichia coli acsP2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol. Microbiol. 51: 241– 254 - PubMed
-
- Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2: 57– 65 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

