Evidence of an evolutionarily conserved LMBR1 domain-containing protein that associates with endocytic cups and plays a role in cell migration in dictyostelium discoideum
- PMID: 22307974
- PMCID: PMC3318307
- DOI: 10.1128/EC.05186-11
Evidence of an evolutionarily conserved LMBR1 domain-containing protein that associates with endocytic cups and plays a role in cell migration in dictyostelium discoideum
Abstract
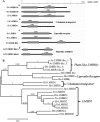
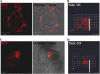
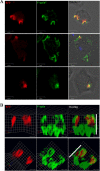
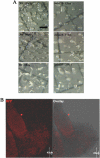
The ampA gene plays a role in Dictyostelium discoideum cell migration. Loss of ampA function results in reduced ability of growing cells to migrate to folic acid and causes small plaques on bacterial lawns, while overexpression of AmpA results in a rapid-migration phenotype and correspondingly larger plaques than seen with wild-type cells. To help understand how the ampA gene functions, second-site suppressors were created by restriction enzyme-mediated integration (REMI) mutagenesis. These mutants were selected for their ability to reduce the large plaque size of the AmpA overexpresser strain. The lmbd2B gene was identified as a suppressor of an AmpA-overexpressing strain. The lmbd2B gene product belongs to the evolutionarily conserved LMBR1 protein family, some of whose known members are endocytic receptors associated with human diseases, such as anemia. In order to understand lmbd2B function, mRFP fusion proteins were created and lmbd2B knockout cell lines were established. Our findings indicate that the LMBD2B protein is found associated with endocytic cups. It colocalizes with proteins that play key roles in endocytic events and is localized to ruffles on the dorsal surfaces of growing cells. Vegetative lmbd2B-null cells display defects in cell migration. These cells have difficulty sensing the chemoattractant folic acid, as indicated by a decrease in their chemotactic index. lmbd2B-null cells also appear to have difficulty establishing a front/back orientation to facilitate migration. A role for lmbd2B in development is also suggested. Our research gives insight into the function of a previously uncharacterized branch of the LMBR1 family of proteins. We provide evidence of an LMBR1 family plasma membrane protein that associates with endocytic cups and plays a role in chemotaxis.
Figures







References
-
- Barlow AL, Macleod A, Noppen S, Sanderson J, Guérin CJ. 2010. Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of Pearson's correlation coefficient. Microsc. Microanal. 16:710–724 - PubMed
-
- Blumberg DD, Ho HN, Petty CL, Varney TR, Gandham S. 2002. AmpA, a modular protein containing disintegrin and ornatin domains, has multiple effects on cell adhesion and cell fate specification. J. Muscle Res. Cell Motil. 23:817–828 - PubMed
-
- Bonner JT. 1967. The cellular slime molds. Princeton University Press, Princeton, NJ
-
- Brazill DT, Meyer LR, Hatton RD, Brock DA, Gomer RH. 2001. ABC transporters required for endocytosis and endosomal pH regulation in Dictyostelium. J. Cell Sci. 114:3923–3932 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

