Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy
- PMID: 22308029
- PMCID: PMC3323008
- DOI: 10.1074/jbc.M111.299917
Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy
Abstract
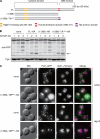
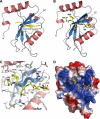
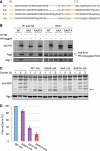
Autophagy-related degradation selective for mitochondria (mitophagy) is an evolutionarily conserved process that is thought to be critical for mitochondrial quality and quantity control. In budding yeast, autophagy-related protein 32 (Atg32) is inserted into the outer membrane of mitochondria with its N- and C-terminal domains exposed to the cytosol and mitochondrial intermembrane space, respectively, and plays an essential role in mitophagy. Atg32 interacts with Atg8, a ubiquitin-like protein localized to the autophagosome, and Atg11, a scaffold protein required for selective autophagy-related pathways, although the significance of these interactions remains elusive. In addition, whether Atg32 is the sole protein necessary and sufficient for initiation of autophagosome formation has not been addressed. Here we show that the Atg32 IMS domain is dispensable for mitophagy. Notably, when anchored to peroxisomes, the Atg32 cytosol domain promoted autophagy-dependent peroxisome degradation, suggesting that Atg32 contains a module compatible for other organelle autophagy. X-ray crystallography reveals that the Atg32 Atg8 family-interacting motif peptide binds Atg8 in a conserved manner. Mutations in this binding interface impair association of Atg32 with the free form of Atg8 and mitophagy. Moreover, Atg32 variants, which do not stably interact with Atg11, are strongly defective in mitochondrial degradation. Finally, we demonstrate that Atg32 forms a complex with Atg8 and Atg11 prior to and independent of isolation membrane generation and subsequent autophagosome formation. Taken together, our data implicate Atg32 as a bipartite platform recruiting Atg8 and Atg11 to the mitochondrial surface and forming an initiator complex crucial for mitophagy.
Figures


References
-
- Weidberg H., Shvets E., Elazar Z. (2011) Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 - PubMed
-
- Mizushima N., Yoshimori T., Ohsumi Y. (2011) Annu. Rev. Cell Dev. Biol. 27, 107–132 - PubMed
-
- Okamoto K., Kondo-Okamoto N. (2012) Biochim. Biophys. Acta in press - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

