Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis
- PMID: 22308031
- PMCID: PMC3323044
- DOI: 10.1074/jbc.M111.318121
Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis
Abstract
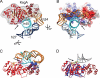
The assembly of the ribosomal subunits is facilitated by ribosome biogenesis factors. The universally conserved methyltransferase KsgA modifies two adjacent adenosine residues in the 3'-terminal helix 45 of the 16 S ribosomal RNA (rRNA). KsgA recognizes its substrate adenosine residues only in the context of a near mature 30S subunit and is required for the efficient processing of the rRNA termini during ribosome biogenesis. Here, we present the cryo-EM structure of KsgA bound to a nonmethylated 30S ribosomal subunit. The structure reveals that KsgA binds to the 30S platform with the catalytic N-terminal domain interacting with substrate adenosine residues in helix 45 and the C-terminal domain making extensive contacts to helix 27 and helix 24. KsgA excludes the penultimate rRNA helix 44 from adopting its position in the mature 30S subunit, blocking the formation of the decoding site and subunit joining. We suggest that the activation of methyltransferase activity and subsequent dissociation of KsgA control conformational changes in helix 44 required for final rRNA processing and translation initiation.
Figures




References
-
- Williamson J. R. (2005) Assembly of the 30S ribosomal subunit. Q. Rev. Biophys. 38, 397–403 - PubMed
-
- Wimberly B. T., Brodersen D. E., Clemons W. M., Jr., Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. (2000) Structure of the 30S ribosomal subunit. Nature 407, 327–339 - PubMed
-
- Mizushima S., Nomura M. (1970) Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226, 1214. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

