Selenocysteine insertion sequence (SECIS)-binding protein 2 alters conformational dynamics of residues involved in tRNA accommodation in 80 S ribosomes
- PMID: 22308032
- PMCID: PMC3323001
- DOI: 10.1074/jbc.M111.320929
Selenocysteine insertion sequence (SECIS)-binding protein 2 alters conformational dynamics of residues involved in tRNA accommodation in 80 S ribosomes
Abstract
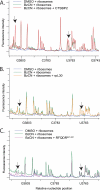
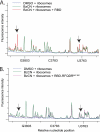
Sec-tRNA(Sec) is site-specifically delivered at defined UGA codons in selenoprotein mRNAs. This recoding event is specified by the selenocysteine insertion sequence (SECIS) element and requires the selenocysteine (Sec)-specific elongation factor, eEFSec, and the SECIS binding protein, SBP2. Sec-tRNA(Sec) is delivered to the ribosome by eEFSec-GTP, but this ternary complex is not sufficient for Sec incorporation, indicating that its access to the ribosomal A-site is regulated. SBP2 stably associates with ribosomes, and mutagenic analysis indicates that this interaction is essential for Sec incorporation. However, the ribosomal function of SBP2 has not been elucidated. To shed light on the functional relevance of the SBP2-ribosome interaction, we screened the functional centers of the 28 S rRNA in translationally competent 80 S ribosomes using selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE). We demonstrate that SBP2 specifically alters the reactivity of specific residues in Helix 89 (H89) and expansion segment 31 (ES31). These results are indicative of a conformational change in response to SBP2 binding. Based on the known functions of H89 during translation, we propose that SBP2 allows Sec incorporation by either promoting Sec-tRNA(Sec) accommodation into the peptidyltransferase center and/or by stimulating the ribosome-dependent GTPase activity of eEFSec.
Figures



References
-
- Diamond A., Dudock B., Hatfield D. (1981) Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell 25, 497–506 - PubMed
-
- Jung J. E., Karoor V., Sandbaken M. G., Lee B. J., Ohama T., Gesteland R. F., Atkins J. F., Mullenbach G. T., Hill K. E., Wahba A. J. (1994) Utilization of selenocysteyl-tRNA[Ser]Sec and seryl-tRNA[Ser]Sec in protein synthesis. J. Biol. Chem. 269, 29739–29745 - PubMed
-
- Fahlman R. P., Dale T., Uhlenbeck O. C. (2004) Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell 16, 799–805 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

