Insights into cis-autoproteolysis reveal a reactive state formed through conformational rearrangement
- PMID: 22308359
- PMCID: PMC3289335
- DOI: 10.1073/pnas.1113633109
Insights into cis-autoproteolysis reveal a reactive state formed through conformational rearrangement
Abstract
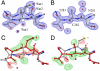
ThnT is a pantetheine hydrolase from the DmpA/OAT superfamily involved in the biosynthesis of the β-lactam antibiotic thienamycin. We performed a structural and mechanistic investigation into the cis-autoproteolytic activation of ThnT, a process that has not previously been subject to analysis within this superfamily of enzymes. Removal of the γ-methyl of the threonine nucleophile resulted in a rate deceleration that we attribute to a reduction in the population of the reactive rotamer. This phenomenon is broadly applicable and constitutes a rationale for the evolutionary selection of threonine nucleophiles in autoproteolytic systems. Conservative substitution of the nucleophile (T282C) allowed determination of a 1.6-Å proenzyme ThnT crystal structure, which revealed a level of structural flexibility not previously observed within an autoprocessing active site. We assigned the major conformer as a nonreactive state that is unable to populate a reactive rotamer. Our analysis shows the system is activated by a structural rearrangement that places the scissile amide into an oxyanion hole and forces the nucleophilic residue into a forbidden region of Ramachandran space. We propose that conformational strain may drive autoprocessing through the destabilization of nonproductive states. Comparison of our data with previous reports uncovered evidence that many inactivated structures display nonreactive conformations. For penicillin and cephalosporin acylases, this discrepancy between structure and function may be resolved by invoking the presence of a hidden conformational state, similar to that reported here for ThnT.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
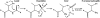


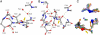
Similar articles
-
Autoproteolytic activation of ThnT results in structural reorganization necessary for substrate binding and catalysis.J Mol Biol. 2012 Sep 28;422(4):508-18. doi: 10.1016/j.jmb.2012.06.012. Epub 2012 Jun 15. J Mol Biol. 2012. PMID: 22706025 Free PMC article.
-
Exploring the role of conformational heterogeneity in cis-autoproteolytic activation of ThnT.Biochemistry. 2014 Jul 8;53(26):4273-81. doi: 10.1021/bi500385d. Epub 2014 Jun 26. Biochemistry. 2014. PMID: 24933323 Free PMC article.
-
Autoproteolytic and catalytic mechanisms for the β-aminopeptidase BapA--a member of the Ntn hydrolase family.Structure. 2012 Nov 7;20(11):1850-60. doi: 10.1016/j.str.2012.07.017. Epub 2012 Sep 12. Structure. 2012. PMID: 22980995
-
Bacterial β-aminopeptidases: structural insights and applications for biocatalysis.Chem Biodivers. 2012 Nov;9(11):2388-409. doi: 10.1002/cbdv.201200305. Chem Biodivers. 2012. PMID: 23161625 Review.
-
Structural and catalytic diversity within the amidohydrolase superfamily.Biochemistry. 2005 May 3;44(17):6383-91. doi: 10.1021/bi047326v. Biochemistry. 2005. PMID: 15850372 Review.
Cited by
-
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.PLoS Pathog. 2014 Jul 24;10(7):e1004228. doi: 10.1371/journal.ppat.1004228. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25058163 Free PMC article.
-
Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.Acta Crystallogr D Struct Biol. 2022 Jul 1;78(Pt 7):911-926. doi: 10.1107/S2059798322005691. Epub 2022 Jun 28. Acta Crystallogr D Struct Biol. 2022. PMID: 35775990 Free PMC article.
-
The Human Ntn-Hydrolase Superfamily: Structure, Functions and Perspectives.Cells. 2022 May 10;11(10):1592. doi: 10.3390/cells11101592. Cells. 2022. PMID: 35626629 Free PMC article. Review.
-
Uncoupling intramolecular processing and substrate hydrolysis in the N-terminal nucleophile hydrolase hASRGL1 by circular permutation.ACS Chem Biol. 2012 Nov 16;7(11):1840-7. doi: 10.1021/cb300232n. Epub 2012 Aug 29. ACS Chem Biol. 2012. PMID: 22891768 Free PMC article.
-
The structure of the PanD/PanZ protein complex reveals negative feedback regulation of pantothenate biosynthesis by coenzyme A.Chem Biol. 2015 Apr 23;22(4):492-503. doi: 10.1016/j.chembiol.2015.03.017. Chem Biol. 2015. PMID: 25910242 Free PMC article.
References
-
- Perler FB, Xu MQ, Paulus H. Protein splicing and autoproteolysis mechanisms. Curr Opin Chem Biol. 1997;1:292–299. - PubMed
-
- Macao B, Johansson DGA, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–76. - PubMed
-
- Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. - PubMed
-
- Lin HH, et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

