Entropic origin of Mg2+-facilitated RNA folding
- PMID: 22308376
- PMCID: PMC3286938
- DOI: 10.1073/pnas.1114859109
Entropic origin of Mg2+-facilitated RNA folding
Abstract
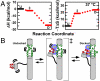
Mg(2+) is essential for the proper folding and function of RNA, though the effect of Mg(2+) concentration on the free energy, enthalpy, and entropy landscapes of RNA folding is unknown. This work exploits temperature-controlled single-molecule FRET methods to address the thermodynamics of RNA folding pathways by probing the intramolecular docking/undocking kinetics of the ubiquitous GAAA tetraloop-receptor tertiary interaction as a function of [Mg(2+)]. These measurements yield the barrier and standard state enthalpies, entropies, and free energies for an RNA tertiary transition, in particular, revealing the thermodynamic origin of [Mg(2+)]-facilitated folding. Surprisingly, these studies reveal that increasing [Mg(2+)] promotes tetraloop-receptor interaction by reducing the entropic barrier (-TΔS(++)(dock)) and the overall entropic penalty (-TΔS(+) (dock)) for docking, with essentially negligible effects on both the activation enthalpy (ΔH(++)(dock)) and overall exothermicity (ΔH(+)(dock)). These observations contrast with the conventional notion that increasing [Mg(2+)] facilitates folding by minimizing electrostatic repulsion of opposing RNA helices, which would incorrectly predict a decrease in ΔH(++)(dock)) and ΔH(+)(dock)) with [Mg(2+)]. Instead we propose that higher [Mg(2+)] can aid RNA folding by decreasing the entropic penalty of counterion uptake and by reducing disorder of the unfolded conformational ensemble.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

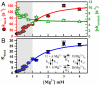
 . Fits of the kdock and kundock titrations with the detailed balance constraint that
. Fits of the kdock and kundock titrations with the detailed balance constraint that  , yield n = 1.8 ± 0.2, k1 = 12.6 ± 0.9 s-1, k2 = 156 ± 23 s-1, k-1 = 8.6 ± 0.7 s-1, k-2 = 5.4 ± 0.2 s-1, Kd = 1.3 ± 0.3 mM, and
, yield n = 1.8 ± 0.2, k1 = 12.6 ± 0.9 s-1, k2 = 156 ± 23 s-1, k-1 = 8.6 ± 0.7 s-1, k-2 = 5.4 ± 0.2 s-1, Kd = 1.3 ± 0.3 mM, and  .
.
 ) and entropy (
) and entropy ( ) of docking from Eq. 1 (Table 1, top, 100 mM NaCl).
) of docking from Eq. 1 (Table 1, top, 100 mM NaCl).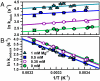
Similar articles
-
Thermodynamic origins of monovalent facilitated RNA folding.Biochemistry. 2012 May 8;51(18):3732-43. doi: 10.1021/bi201420a. Epub 2012 Apr 23. Biochemistry. 2012. PMID: 22448852 Free PMC article.
-
Enthalpy-driven RNA folding: single-molecule thermodynamics of tetraloop-receptor tertiary interaction.Biochemistry. 2009 Mar 24;48(11):2550-8. doi: 10.1021/bi8019788. Biochemistry. 2009. PMID: 19186984 Free PMC article.
-
The role of counterion valence and size in GAAA tetraloop-receptor docking/undocking kinetics.J Mol Biol. 2012 Oct 19;423(2):198-216. doi: 10.1016/j.jmb.2012.07.006. Epub 2012 Jul 14. J Mol Biol. 2012. PMID: 22796627
-
An RNA folding motif: GNRA tetraloop-receptor interactions.Q Rev Biophys. 2013 Aug;46(3):223-64. doi: 10.1017/S0033583513000048. Epub 2013 Aug 5. Q Rev Biophys. 2013. PMID: 23915736 Review.
-
Probing the kinetic and thermodynamic consequences of the tetraloop/tetraloop receptor monovalent ion-binding site in P4-P6 RNA by smFRET.Biochem Soc Trans. 2015 Apr;43(2):172-8. doi: 10.1042/BST20140268. Biochem Soc Trans. 2015. PMID: 25849913 Free PMC article. Review.
Cited by
-
Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites.Nucleic Acids Res. 2014 Feb;42(4):2646-59. doi: 10.1093/nar/gkt1139. Epub 2013 Nov 14. Nucleic Acids Res. 2014. PMID: 24234441 Free PMC article.
-
Single-molecule fluorescence resonance energy transfer studies of the human telomerase RNA pseudoknot: temperature-/urea-dependent folding kinetics and thermodynamics.J Phys Chem B. 2014 Apr 10;118(14):3853-63. doi: 10.1021/jp501893c. Epub 2014 Mar 28. J Phys Chem B. 2014. PMID: 24617561 Free PMC article.
-
Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics.Proc Natl Acad Sci U S A. 2014 Jun 10;111(23):8464-9. doi: 10.1073/pnas.1316039111. Epub 2014 May 21. Proc Natl Acad Sci U S A. 2014. PMID: 24850865 Free PMC article.
-
Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine-Specific Cancer Targeting with Undetectable Toxicity.Chem Rev. 2021 Jul 14;121(13):7398-7467. doi: 10.1021/acs.chemrev.1c00009. Epub 2021 May 26. Chem Rev. 2021. PMID: 34038115 Free PMC article. Review.
-
From static to dynamic: the need for structural ensembles and a predictive model of RNA folding and function.Curr Opin Struct Biol. 2015 Feb;30:125-133. doi: 10.1016/j.sbi.2015.02.006. Epub 2015 Mar 2. Curr Opin Struct Biol. 2015. PMID: 25744941 Free PMC article. Review.
References
-
- Tinoco I, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. - PubMed
-
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomolec Struct. 1997;26:113–137. - PubMed
-
- Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol. 2006;16:270–278. - PubMed
-
- Zhuang XW, et al. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

