Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora
- PMID: 22308384
- PMCID: PMC3289388
- DOI: 10.1073/pnas.1115346109
Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora
Abstract
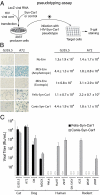
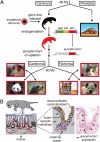
Syncytins are envelope protein genes of retroviral origin that have been captured for a function in placentation. Two such genes have already been identified in simians, two distinct, unrelated genes have been identified in Muridae, and a fifth gene has been identified in the rabbit. Here, we searched for similar genes in the Laurasiatheria clade, which diverged from Euarchontoglires--primates, rodents, and lagomorphs--shortly after mammalian radiation (100 Mya). In silico search for envelope protein genes with full-coding capacity within the dog and cat genomes identified several candidate genes, with one common to both species that displayed placenta-specific expression, which was revealed by RT-PCR analysis of a large panel of tissues. This gene belongs to a degenerate endogenous retroviral element, with precise proviral integration at a site common to dog and cat. Cloning of the gene for an ex vivo pseudotype assay showed fusogenicity on both dog and cat cells. In situ hybridization on placenta sections from both species showed specific expression at the level of the invasive fetal villi within the placental junctional zone, where trophoblast cells fuse into a syncytiotrophoblast layer to form the maternofetal interface. Finally, we show that the gene is conserved among a series of 26 Carnivora representatives, with evidence for purifying selection and conservation of fusogenic activity. The gene is not found in the Pholidota order and, therefore, it was captured before Carnivora radiation, between 60 and 85 Mya. This gene is the oldest syncytin gene identified to date, and it is the first in a new major clade of eutherian mammals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Comment in
-
Retroviruses push the envelope for mammalian placentation.Proc Natl Acad Sci U S A. 2012 Feb 14;109(7):2184-5. doi: 10.1073/pnas.1121365109. Epub 2012 Jan 26. Proc Natl Acad Sci U S A. 2012. PMID: 22308481 Free PMC article. No abstract available.
References
-
- Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. - PubMed
-
- Cáceres M, Thomas JW, Thomas JW. NISC Comparative Sequencing Program The gene of retroviral origin Syncytin 1 is specific to hominoids and is inactive in Old World monkeys. J Hered. 2006;97:100–106. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

