Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development
- PMID: 22308433
- PMCID: PMC3289350
- DOI: 10.1073/pnas.1119112109
Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development
Abstract
Understanding the mechanisms of epigenetic remodeling that follow fertilization is a fundamental step toward understanding the bases of early embryonic development and pluripotency. Extensive and dynamic chromatin remodeling is observed after fertilization, including DNA methylation and histone modifications. These changes underlie the transition from gametic to embryonic chromatin and are thought to facilitate embryonic genome activation. In particular, trimethylation of histone 3 lysine 27 (H3K27me3) is associated with gene-specific transcription repression. Global levels of this epigenetic mark are high in oocyte chromatin and decrease to minimal levels at the time of embryonic genome activation. We provide evidence that the decrease in H3K27me3 observed during early development is cell-cycle independent, suggesting an active mechanism for removal of this epigenetic mark. Among H3K27me3-specific demethylases, Jumonji domain-containing protein 3 (JMJD3), but not ubiquitously transcribed tetratricopeptide repeat X (UTX), present high transcript levels in oocytes. Soon after fertilization JMJD3 protein levels increase, concurrent with a decrease in mRNA levels. This pattern of expression suggests maternal inheritance of JMJD3. Knockdown of JMJD3 by siRNA injection in parthenogenetically activated metaphase II oocytes resulted in inhibition of the H3K27me3 decrease normally observed in preimplantation embryos. Moreover, knockdown of JMJD3 in oocytes reduced the rate of blastocyst development. Overall, these results indicate that JMJD3 is involved in active demethylation of H3K27me3 during early embryo development and that this mark plays an important role during the progression of embryos to blastocysts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



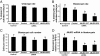
References
-
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. - PubMed
-
- Worrad DM, Turner BM, Schultz RM. Temporally restricted spatial localization of acetylated isoforms of histone H4 and RNA polymerase II in the 2-cell mouse embryo. Development. 1995;121:2949–2959. - PubMed
-
- Braude PR, Monk M, Pickering SJ, Cant A, Johnson MH. Measurement of HPRT activity in the human unfertilized oocyte and pre-embryo. Prenat Diagn. 1989;9:839–850. - PubMed
-
- Memili E, First NL. Developmental changes in RNA polymerase II in bovine oocytes, early embryos, and effect of alpha-amanitin on embryo development. Mol Reprod Dev. 1998;51:381–389. - PubMed
-
- Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15:531–538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

