Quantification and visualization of phosphoinositides by quantum dot-labeled specific binding-domain probes
- PMID: 22308508
- PMCID: PMC3307658
- DOI: 10.1194/jlr.D019547
Quantification and visualization of phosphoinositides by quantum dot-labeled specific binding-domain probes
Abstract
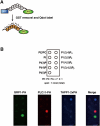
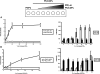
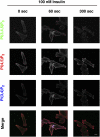
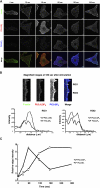
Phosphoinositides (PI) play important regulatory roles in cell physiology. Localization and quantitation of PIs within the cell is necessary to understand their precise function. Currently, ectopic expression of green fluorescent protein (GFP)-fused PI-binding domains is used to visualize PIs localized to the cell membrane. However, ectopically expressed PI-binding domains may compete with endogenous binding proteins, thus altering the physiological functions of the PIs. Here, we establish a novel method for quantification and visualization of PIs in cells and tissue samples using PI-binding domains labeled with quantum dots (Qdot) as specific probes. This method allowed us to simultaneously quantify three distinct PIs, phosphatidylinositol 3,4,5-triphosphatase [PtdIns(3,4,5)P(3)), PtdIns(3,4)P(2), and PtdIns(4,5)P(2), in crude acidic lipids extracted from insulin-stimulated cells. In addition, the method allowed the PIs to be visualized within fixed cells and tissues. Sequential and spatial changes in PI production and distribution were detected in platelet-derived growth factor (PDGF)-stimulated NRK49F cells. We also observed accumulation of PtdIns(3,4)P(2) at the dorsal ruffle in PDGF-stimulated NIH3T3 cells. Finally, we found PtdIns(3,4,5)P(3) was enriched in lung cancer tissues, which also showed high levels of phosphorylated Akt. Our new method to quantify and visualize PIs is expected to provide further insight into the role of lipid signaling in a wide range of cellular events.
Figures




References
-
- Janmey P. A., Lindberg U. 2004. Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cell Biol. 5: 658–666 - PubMed
-
- Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443: 651–657 - PubMed
-
- Lazar D. F., Saltiel A. R. 2006. Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat. Rev. Drug Discov. 5: 333–342 - PubMed
-
- Ooms L. M., Horan K. A., Rahman P., Seaton G., Gurung R., Kethesparan D. S., Mitchell C. A. 2009. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 419: 29–49 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

