Lectin chromatography/mass spectrometry discovery workflow identifies putative biomarkers of aggressive breast cancers
- PMID: 22309216
- PMCID: PMC3383053
- DOI: 10.1021/pr201206w
Lectin chromatography/mass spectrometry discovery workflow identifies putative biomarkers of aggressive breast cancers
Abstract
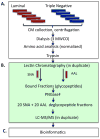
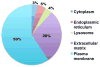
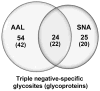
We used a lectin chromatography/MS-based approach to screen conditioned medium from a panel of luminal (less aggressive) and triple negative (more aggressive) breast cancer cell lines (n=5/subtype). The samples were fractionated using the lectins Aleuria aurantia (AAL) and Sambucus nigra agglutinin (SNA), which recognize fucose and sialic acid, respectively. The bound fractions were enzymatically N-deglycosylated and analyzed by LC-MS/MS. In total, we identified 533 glycoproteins, ∼90% of which were components of the cell surface or extracellular matrix. We observed 1011 glycosites, 100 of which were solely detected in ≥3 triple negative lines. Statistical analyses suggested that a number of these glycosites were triple negative-specific and thus potential biomarkers for this tumor subtype. An analysis of RNaseq data revealed that approximately half of the mRNAs encoding the protein scaffolds that carried potential biomarker glycosites were up-regulated in triple negative vs luminal cell lines, and that a number of genes encoding fucosyl- or sialyltransferases were differentially expressed between the two subtypes, suggesting that alterations in glycosylation may also drive candidate identification. Notably, the glycoproteins from which these putative biomarker candidates were derived are involved in cancer-related processes. Thus, they may represent novel therapeutic targets for this aggressive tumor subtype.
Figures



References
-
- Clowers BH, Dodds ED, Seipert RR, Lebrilla CB. Site determination of protein glycosylation based on digestion with immobilized nonspecific proteases and Fourier transform ion cyclotron resonance mass spectrometry. J Proteome Res. 2007;6(10):4032–40. - PubMed
-
- Duffy MJ, Evoy D, McDermott EW. CA 15–3: uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411(23–24):1869–74. - PubMed
-
- Orntoft TF, Vestergaard EM. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis. 1999;20(2):362–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

