Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter
- PMID: 22309491
- PMCID: PMC3293747
- DOI: 10.1186/1471-2164-13-62
Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter
Abstract
Background: Aspergillus fumigatus is a mold responsible for the majority of cases of aspergillosis in humans. To survive in the human body, A. fumigatus must adapt to microenvironments that are often characterized by low nutrient and oxygen availability. Recent research suggests that the ability of A. fumigatus and other pathogenic fungi to adapt to hypoxia contributes to their virulence. However, molecular mechanisms of A. fumigatus hypoxia adaptation are poorly understood. Thus, to better understand how A. fumigatus adapts to hypoxic microenvironments found in vivo during human fungal pathogenesis, the dynamic changes of the fungal transcriptome and proteome in hypoxia were investigated over a period of 24 hours utilizing an oxygen-controlled fermenter system.
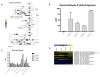
Results: Significant increases in transcripts associated with iron and sterol metabolism, the cell wall, the GABA shunt, and transcriptional regulators were observed in response to hypoxia. A concomitant reduction in transcripts was observed with ribosome and terpenoid backbone biosynthesis, TCA cycle, amino acid metabolism and RNA degradation. Analysis of changes in transcription factor mRNA abundance shows that hypoxia induces significant positive and negative changes that may be important for regulating the hypoxia response in this pathogenic mold. Growth in hypoxia resulted in changes in the protein levels of several glycolytic enzymes, but these changes were not always reflected by the corresponding transcriptional profiling data. However, a good correlation overall (R(2) = 0.2, p < 0.05) existed between the transcriptomic and proteomics datasets for all time points. The lack of correlation between some transcript levels and their subsequent protein levels suggests another regulatory layer of the hypoxia response in A. fumigatus.
Conclusions: Taken together, our data suggest a robust cellular response that is likely regulated both at the transcriptional and post-transcriptional level in response to hypoxia by the human pathogenic mold A. fumigatus. As with other pathogenic fungi, the induction of glycolysis and transcriptional down-regulation of the TCA cycle and oxidative phosphorylation appear to major components of the hypoxia response in this pathogenic mold. In addition, a significant induction of the transcripts involved in ergosterol biosynthesis is consistent with previous observations in the pathogenic yeasts Candida albicans and Cryptococcus neoformans indicating conservation of this response to hypoxia in pathogenic fungi. Because ergosterol biosynthesis enzymes also require iron as a co-factor, the increase in iron uptake transcripts is consistent with an increased need for iron under hypoxia. However, unlike C. albicans and C. neoformans, the GABA shunt appears to play an important role in reducing NADH levels in response to hypoxia in A. fumigatus and it will be intriguing to determine whether this is critical for fungal virulence. Overall, regulatory mechanisms of the A. fumigatus hypoxia response appear to involve both transcriptional and post-transcriptional control of transcript and protein levels and thus provide candidate genes for future analysis of their role in hypoxia adaptation and fungal virulence.
Figures









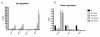


References
-
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. - PubMed
-
- Gurcan S, Tikvesli M, Eryildiz C, Evci C, Ener B. Antifungal susceptibilities of Aspergillus spp. strains isolated from invasive aspergillosis cases. Mikrobiyoloji Bulteni. 2010;44:273–278. - PubMed
-
- Barron MA, Madinger NE. Opportunistic fungal infections, part 2: Candida and Aspergillus. Infect Med. 2008;25:498–505.
-
- Howard SJ, Pasqualotto AC, Denning DW. Azole resistance in allergic bronchopulmonary aspergillosis and Aspergillus bronchitis. Clin Microbiol Infect. 2010;16:683–688. - PubMed
-
- Bonatti H, Lass-Floerl C, Angerer K, Singh N, Lechner M, Stelzmueller I, Singh R, Schmid T, Geltner C. Successful management of postpneumonectomy Aspergillus pleural empyema by combined surgical and anti-fungal treatment with voriconazole and caspofungin. Mycoses. 2010;53:448–454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

