Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells
- PMID: 22310368
- PMCID: PMC3314980
- DOI: 10.1016/j.mito.2012.01.001
Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells
Abstract
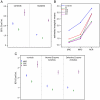
Given the complexity of the respiratory chain structure, assembly and regulation, the diagnostic workout for the identification of defects of oxidative phosphorylation (OXPHOS) is a major challenge. Spectrophotometric assays, that measure the activity of individual respiratory complexes in tissue and cell homogenates or isolated mitochondria, are highly specific, but their utilization is limited by the availability of sufficient biological material and intrinsic sensitivity. A further limitation is tissue specificity, which usually determines attenuation, or disappearance, in cultured fibroblasts, of defects detected in muscle or liver. We used numerous fibroblast cell lines derived from patients with OXPHOS deficiencies to set up experimental protocols required for the direct readout of cellular respiration using the Seahorse XF96 apparatus, which measures oxygen consumption rate (OCR) and extra-cellular acidification rate (ECAR) in 96 well plates. Results demonstrate that first level screening based on microscale oxygraphy is more sensitive, cheaper and rapid than spectrophotometry for the biochemical evaluation of cells from patients with suspected mitochondrial disorders.
Copyright © 2012 Elsevier B.V. and Mitochondria Research Society. All rights reserved. All rights reserved.
Figures



References
-
- Acín-Pérez R., Fernández-Silva P., Peleato M.L., Pérez-Martos A., Enriquez J.A. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. - PubMed
-
- Benit P., Goncalves S., Philippe D.E., Briere J.J., Martin G., Rustin P. Three spectrophotometric assays for the measurements of the five respiratory chain complexes in minuscule biological samples. Clin. Chim. Acta. 2006;34:283–292. - PubMed
-
- Bugiani M., Invernizzi F., Alberio S., Briem E., Lamantea E., Carrara F., Moroni I., Farina L., Spada M., Donati M.A., Uziel G., Zeviani M. Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta. 2004;1659:136–147. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

