Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules
- PMID: 22312047
- PMCID: PMC3267113
- DOI: 10.1098/rstb.2011.0266
Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules
Abstract
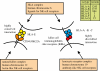
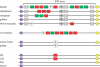
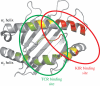
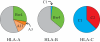
In placental mammals, natural killer (NK) cells are a population of lymphocytes that make unique contributions to immune defence and reproduction, functions essential for survival of individuals, populations and species. Modulating these functions are conserved and variable NK-cell receptors that recognize epitopes of major histocompatibility complex (MHC) class I molecules. In humans, for example, recognition of human leucocyte antigen (HLA)-E by the CD94:NKG2A receptor is conserved, whereas recognition of HLA-A, B and C by the killer cell immunoglobulin-like receptors (KIRs) is diversified. Competing demands of the immune and reproductive systems, and of T-cell and NK-cell immunity-combined with the segregation on different chromosomes of variable NK-cell receptors and their MHC class I ligands-drive an unusually rapid evolution that has resulted in unprecedented levels of species specificity, as first appreciated from comparison of mice and humans. Counterparts to human KIR are present only in simian primates. Observed in these species is the coevolution of KIR and the four MHC class I epitopes to which human KIR recognition is restricted. Unique to hominids is the emergence of the MHC-C locus as a supplier of specialized and superior ligands for KIR. This evolutionary trend is most highly elaborated in the chimpanzee. Unique to the human KIR locus are two groups of KIR haplotypes that are present in all human populations and subject to balancing selection. Group A KIR haplotypes resemble chimpanzee KIR haplotypes and are enriched for genes encoding KIR that bind HLA class I, whereas group B KIR haplotypes are enriched for genes encoding receptors with diminished capacity to bind HLA class I. Correlating with their balance in human populations, B haplotypes favour reproductive success, whereas A haplotypes favour successful immune defence. Evolution of the B KIR haplotypes is thus unique to the human species.
Figures





References
-
- Parham P. 2005. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5, 201–21410.1038/nri1570 (doi:10.1038/nri1570) - DOI - DOI - PubMed
-
- Vivier E., Raulet D. H., Moretta A., Caligiuri M. A., Zitvogel L., Lanier L. L., Yokoyama W. M., Ugolini S. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–4910.1126/science.1198687 (doi:10.1126/science.1198687) - DOI - DOI - PMC - PubMed
-
- Moffett A., Loke C. 2006. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 6, 584–59410.1038/nri1897 (doi:10.1038/nri1897) - DOI - DOI - PubMed
-
- Anfossi N., et al. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25, 331–34210.1016/j.immuni.2006.06.013 (doi:10.1016/j.immuni.2006.06.013) - DOI - DOI - PubMed
-
- Yawata M., Yawata N., Draghi M., Partheniou F., Little A. M., Parham P. 2008. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood 112, 2369–238010.1182/blood-2008-03-143727 (doi:10.1182/blood-2008-03-143727) - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
