Near-full genome characterisation of two natural intergenotypic 2k/1b recombinant hepatitis C virus isolates
- PMID: 22315602
- PMCID: PMC3270303
- DOI: 10.1155/2011/710438
Near-full genome characterisation of two natural intergenotypic 2k/1b recombinant hepatitis C virus isolates
Abstract
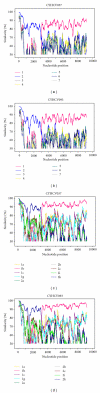
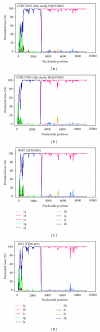
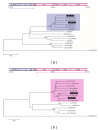
Few natural intergenotypic hepatitis C virus (HCV) recombinants have been characterised, and only RF1_2k/1b has demonstrated widespread transmission. The near-full length genome sequences for two cases of 2k/1b recombinants (CYHCV037 and CYHCV093) sampled in Cyprus were obtained using strain-specific RT-PCR amplification and sequencing protocols. Sequence analysis confirmed their similarity with the original RF1_2k/1b strain from St. Petersburg, N687. These two isolates significantly contribute to the sequence data available on this recombinant and confirm its increasing spread among individuals from Eastern Europe, and its association with transmission through intravenous drug use. Phylogenetic analyses reveal clustering of the sequence 3' to the recombination point, not seen in the topology of the 5' sequences, implying a more complicated evolutionary history than that held to date. The increasing cases of HCV recombinant strains underline the requirement of their contribution to the standardised rules of HCV classification and nomenclature, molecular epidemiology, diagnosis, and treatment.
Figures
References
-
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. - PubMed
-
- Simmonds P. Genetic diversity and evolution of hepatitis C virus—15 years on. Journal of General Virology. 2004;85(11):3173–3188. - PubMed
-
- Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–973. - PubMed
-
- Colina R, Casane D, Vasquez S, et al. Evidence of intratypic recombination in natural populations of hepatitis C virus. Journal of General Virology. 2004;85(1):31–37. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

