Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search
- PMID: 22318518
- PMCID: PMC3288143
- DOI: 10.1038/nature10782
Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search
Abstract
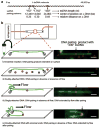
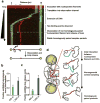
DNA breaks can be repaired with high fidelity by homologous recombination. A ubiquitous protein that is essential for this DNA template-directed repair is RecA. After resection of broken DNA to produce single-stranded DNA (ssDNA), RecA assembles on this ssDNA into a filament with the unique capacity to search and find DNA sequences in double-stranded DNA (dsDNA) that are homologous to the ssDNA. This homology search is vital to recombinational DNA repair, and results in homologous pairing and exchange of DNA strands. Homologous pairing involves DNA sequence-specific target location by the RecA-ssDNA complex. Despite decades of study, the mechanism of this enigmatic search process remains unknown. RecA is a DNA-dependent ATPase, but ATP hydrolysis is not required for DNA pairing and strand exchange, eliminating active search processes. Using dual optical trapping to manipulate DNA, and single-molecule fluorescence microscopy to image DNA pairing, we demonstrate that both the three-dimensional conformational state of the dsDNA target and the length of the homologous RecA-ssDNA filament have important roles in the homology search. We discovered that as the end-to-end distance of the target dsDNA molecule is increased, constraining the available three-dimensional (3D) conformations of the molecule, the rate of homologous pairing decreases. Conversely, when the length of the ssDNA in the nucleoprotein filament is increased, homology is found faster. We propose a model for the DNA homology search process termed 'intersegmental contact sampling', in which the intrinsic multivalent nature of the RecA nucleoprotein filament is used to search DNA sequence space within 3D domains of DNA, exploiting multiple weak contacts to rapidly search for homology. Our findings highlight the importance of the 3D conformational dynamics of DNA, reveal a previously unknown facet of the homology search, and provide insight into the mechanism of DNA target location by this member of a universal family of proteins.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol. 1985;181:281–295. - PubMed
-
- Adzuma K. No sliding during homology search by RecA protein. J Biol Chem. 1998;273:31565–31573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

