PASSEL: the PeptideAtlas SRMexperiment library
- PMID: 22318887
- PMCID: PMC3832291
- DOI: 10.1002/pmic.201100515
PASSEL: the PeptideAtlas SRMexperiment library
Abstract
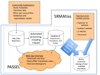
Public repositories for proteomics data have accelerated proteomics research by enabling more efficient cross-analyses of datasets, supporting the creation of protein and peptide compendia of experimental results, supporting the development and testing of new software tools, and facilitating the manuscript review process. The repositories available to date have been designed to accommodate either shotgun experiments or generic proteomic data files. Here, we describe a new kind of proteomic data repository for the collection and representation of data from selected reaction monitoring (SRM) measurements. The PeptideAtlas SRM Experiment Library (PASSEL) allows researchers to easily submit proteomic data sets generated by SRM. The raw data are automatically processed in a uniform manner and the results are stored in a database, where they may be downloaded or browsed via a web interface that includes a chromatogram viewer. PASSELenables cross-analysis of SRMdata, supports optimization of SRMdata collection, and facilitates the review process of SRMdata. Further, PASSELwill help in the assessment of proteotypic peptide performance in a wide array of samples containing the same peptide, as well as across multiple experimental protocols.
© 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors declare no financial or commercial conflicts of interest.
Figures



References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

