Human gene copy number spectra analysis in congenital heart malformations
- PMID: 22318994
- PMCID: PMC3426426
- DOI: 10.1152/physiolgenomics.00013.2012
Human gene copy number spectra analysis in congenital heart malformations
Abstract
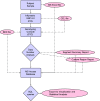
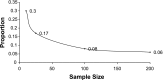
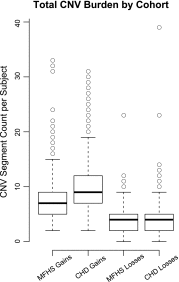
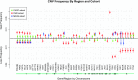
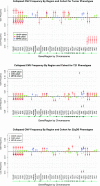
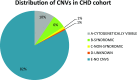
The clinical significance of copy number variants (CNVs) in congenital heart disease (CHD) continues to be a challenge. Although CNVs including genes can confer disease risk, relationships between gene dosage and phenotype are still being defined. Our goal was to perform a quantitative analysis of CNVs involving 100 well-defined CHD risk genes identified through previously published human association studies in subjects with anatomically defined cardiac malformations. A novel analytical approach permitting CNV gene frequency "spectra" to be computed over prespecified regions to determine phenotype-gene dosage relationships was employed. CNVs in subjects with CHD (n = 945), subphenotyped into 40 groups and verified in accordance with the European Paediatric Cardiac Code, were compared with two control groups, a disease-free cohort (n = 2,026) and a population with coronary artery disease (n = 880). Gains (≥200 kb) and losses (≥100 kb) were determined over 100 CHD risk genes and compared using a Barnard exact test. Six subphenotypes showed significant enrichment (P ≤ 0.05), including aortic stenosis (valvar), atrioventricular canal (partial), atrioventricular septal defect with tetralogy of Fallot, subaortic stenosis, tetralogy of Fallot, and truncus arteriosus. Furthermore, CNV gene frequency spectra were enriched (P ≤ 0.05) for losses at: FKBP6, ELN, GTF2IRD1, GATA4, CRKL, TBX1, ATRX, GPC3, BCOR, ZIC3, FLNA and MID1; and gains at: PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, HRAS, GATA6 and RUNX1. Of CHD subjects, 14% had causal chromosomal abnormalities, and 4.3% had likely causal (significantly enriched), large, rare CNVs. CNV frequency spectra combined with precision phenotyping may lead to increased molecular understanding of etiologic pathways.
Figures

References
-
- Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, Lalani SR, Chinault AC, Cheung SW, Lupski JR, Patel A. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet 82: 214–221, 2008. - PMC - PubMed
-
- Bondy CA. Congenital cardiovascular disease in Turner syndrome. Congenit Heart Dis 3: 2–15, 2008. - PubMed
-
- Bondy CA, Cheng C. Monosomy for the X chromosome. Chromosome Res 17: 649–658, 2009. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

