Evidence for widespread positive and purifying selection across the European rabbit (Oryctolagus cuniculus) genome
- PMID: 22319161
- PMCID: PMC3375474
- DOI: 10.1093/molbev/mss025
Evidence for widespread positive and purifying selection across the European rabbit (Oryctolagus cuniculus) genome
Abstract
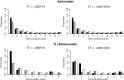
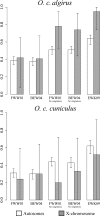
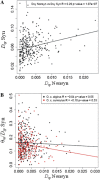
The nearly neutral theory of molecular evolution predicts that the efficacy of both positive and purifying selection is a function of the long-term effective population size (N(e)) of a species. Under this theory, the efficacy of natural selection should increase with N(e). Here, we tested this simple prediction by surveying ~1.5 to 1.8 Mb of protein coding sequence in the two subspecies of the European rabbit (Oryctolagus cuniculus algirus and O. c. cuniculus), a mammal species characterized by high levels of nucleotide diversity and N(e) estimates for each subspecies on the order of 1 × 10(6). When the segregation of slightly deleterious mutations and demographic effects were taken into account, we inferred that >60% of amino acid substitutions on the autosomes were driven to fixation by positive selection. Moreover, we inferred that a small fraction of new amino acid mutations (<4%) are effectively neutral (defined as 0 < N(e)s < 1) and that this fraction was negatively correlated with a gene's expression level. Consistent with models of recurrent adaptive evolution, we detected a negative correlation between levels of synonymous site polymorphism and the rate of protein evolution, although the correlation was weak and nonsignificant. No systematic X chromosome-autosome difference was found in the efficacy of selection. For example, the proportion of adaptive substitutions was significantly higher on the X chromosome compared with the autosomes in O. c. algirus but not in O. c. cuniculus. Our findings support widespread positive and purifying selection in rabbits and add to a growing list of examples suggesting that differences in N(e) among taxa play a substantial role in determining rates and patterns of protein evolution.
Figures
References
-
- Aquadro CF, Bauer DuMont V, Reed FA. Genome-wide variation in the human and fruitfly: a comparison. Curr Opin Genet Dev. 2001;11:627–634. - PubMed
-
- Axelsson E, Ellegren H. Quantification of adaptive evolution of genes expressed in avian brain and the population size effect on the efficacy of selection. Mol Biol Evol. 2009;26:1073–1079. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

