Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress
- PMID: 22321773
- PMCID: PMC3339519
- DOI: 10.1186/1471-2164-13-64
Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress
Abstract
Background: Cassava is an important tropical root crop adapted to a wide range of environmental stimuli such as drought and acid soils. Nevertheless, it is an extremely cold-sensitive tropical species. Thus far, there is limited information about gene regulation and signalling pathways related to the cold stress response in cassava. The development of microarray technology has accelerated the study of global transcription profiling under certain conditions.
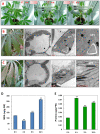
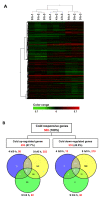
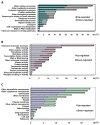
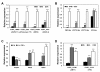
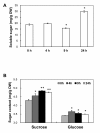
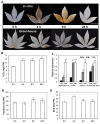
Results: A 60-mer oligonucleotide microarray representing 20,840 genes was used to perform transcriptome profiling in apical shoots of cassava subjected to cold at 7°C for 0, 4 and 9 h. A total of 508 transcripts were identified as early cold-responsive genes in which 319 sequences had functional descriptions when aligned with Arabidopsis proteins. Gene ontology annotation analysis identified many cold-relevant categories, including 'Response to abiotic and biotic stimulus', 'Response to stress', 'Transcription factor activity', and 'Chloroplast'. Various stress-associated genes with a wide range of biological functions were found, such as signal transduction components (e.g., MAP kinase 4), transcription factors (TFs, e.g., RAP2.11), and reactive oxygen species (ROS) scavenging enzymes (e.g., catalase 2), as well as photosynthesis-related genes (e.g., PsaL). Seventeen major TF families including many well-studied members (e.g., AP2-EREBP) were also involved in the early response to cold stress. Meanwhile, KEGG pathway analysis uncovered many important pathways, such as 'Plant hormone signal transduction' and 'Starch and sucrose metabolism'. Furthermore, the expression changes of 32 genes under cold and other abiotic stress conditions were validated by real-time RT-PCR. Importantly, most of the tested stress-responsive genes were primarily expressed in mature leaves, stem cambia, and fibrous roots rather than apical buds and young leaves. As a response to cold stress in cassava, an increase in transcripts and enzyme activities of ROS scavenging genes and the accumulation of total soluble sugars (including sucrose and glucose) were also detected.
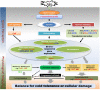
Conclusions: The dynamic expression changes reflect the integrative controlling and transcriptome regulation of the networks in the cold stress response of cassava. The biological processes involved in the signal perception and physiological response might shed light on the molecular mechanisms related to cold tolerance in tropical plants and provide useful candidate genes for genetic improvement.
Figures
References
-
- El-Sharkawy MA. International research on cassava photosynthesis, productivity, eco-physiology, and responses to environmental stresses in the tropics. Photosynthetica. 2006;44:481–512. doi: 10.1007/s11099-006-0063-0. - DOI
-
- Chen QL, Li Z, Fan GZ, Zhu KY, Zhang W, Zhu HQ. Indications of stratospheric anomalies in the freezing rain and snow disaster in South China, 2008. Sci China Earth Sci. 2011;54:1248–1256. doi: 10.1007/s11430-011-4192-3. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

