The global repressor FliZ antagonizes gene expression by σS-containing RNA polymerase due to overlapping DNA binding specificity
- PMID: 22323519
- PMCID: PMC3367168
- DOI: 10.1093/nar/gks055
The global repressor FliZ antagonizes gene expression by σS-containing RNA polymerase due to overlapping DNA binding specificity
Abstract
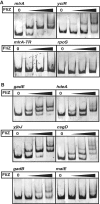
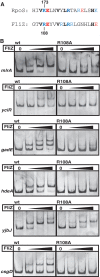
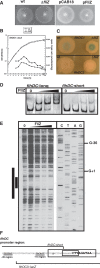
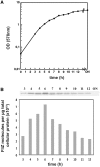
FliZ, a global regulatory protein under the control of the flagellar master regulator FlhDC, was shown to antagonize σ(S)-dependent gene expression in Escherichia coli. Thereby it plays a pivotal role in the decision between alternative life-styles, i.e. FlhDC-controlled flagellum-based motility or σ(S)-dependent curli fimbriae-mediated adhesion and biofilm formation. Here, we show that FliZ is an abundant DNA-binding protein that inhibits gene expression mediated by σ(S) by recognizing operator sequences that resemble the -10 region of σ(S)-dependent promoters. FliZ does so with a structural element that is similar to region 3.0 of σ(S). Within this element, R108 in FliZ corresponds to K173 in σ(S), which contacts a conserved cytosine at the -13 promoter position that is specific for σ(S)-dependent promoters. R108 as well as C(-13) are also crucial for DNA binding by FliZ. However, while a number of FliZ binding sites correspond to known σ(S)-dependent promoters, promoter activity is not a prerequisite for FliZ binding and repressor function. Thus, we demonstrate that FliZ also feedback-controls flagellar gene expression by binding to a site in the flhDC control region that shows similarity only to a -10 element of a σ(S)-dependent promoter, but does not function as a promoter.
Figures



References
-
- Ferenci T. Hungry bacteria-definition and properties of a nutritional state. Environ. Microbiol. 2001;3:605–611. - PubMed
-
- Hengge R. The general stress response in Gram-negative bacteria. In: Storz G, Hengge R, editors. Bacterial Stress Responses. Washington, DC: ASM Press; 2010. pp. 25–289.
-
- Nyström T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 2004;54:855–862. - PubMed
-
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

