Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation
- PMID: 22323818
- PMCID: PMC4693633
- DOI: 10.1126/science.1214453
Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation
Abstract
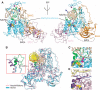
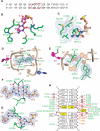
DNMT1, the major maintenance DNA methyltransferase in animals, helps to regulate gene expression, genome imprinting, and X-chromosome inactivation. We report on the crystal structure of a productive covalent mouse DNMT1(731-1602)-DNA complex containing a central hemimethylated CpG site. The methyl group of methylcytosine is positioned within a shallow hydrophobic concave surface, whereas the cytosine on the target strand is looped out and covalently anchored within the catalytic pocket. The DNA is distorted at the hemimethylated CpG step, with side chains from catalytic and recognition loops inserting through both grooves to fill an intercalation-type cavity associated with a dual base flip-out on partner strands. Structural and biochemical data establish how a combination of active and autoinhibitory mechanisms ensures the high fidelity of DNMT1-mediated maintenance DNA methylation.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

