RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3
- PMID: 22325148
- PMCID: PMC3281992
- DOI: 10.1016/j.cell.2011.12.029
RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3
Erratum in
- Cell. 2012 Jun 22;149(7):1647-8
Abstract
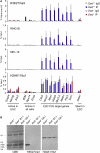
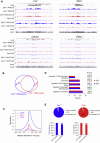
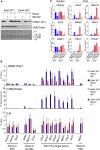
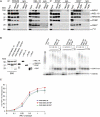
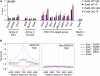
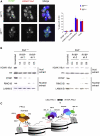
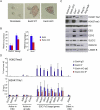
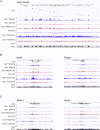
Polycomb-repressive complex 1 (PRC1) has a central role in the regulation of heritable gene silencing during differentiation and development. PRC1 recruitment is generally attributed to interaction of the chromodomain of the core protein Polycomb with trimethyl histone H3K27 (H3K27me3), catalyzed by a second complex, PRC2. Unexpectedly we find that RING1B, the catalytic subunit of PRC1, and associated monoubiquitylation of histone H2A are targeted to closely overlapping sites in wild-type and PRC2-deficient mouse embryonic stem cells (mESCs), demonstrating an H3K27me3-independent pathway for recruitment of PRC1 activity. We show that this pathway is mediated by RYBP-PRC1, a complex comprising catalytic subunits of PRC1 and the protein RYBP. RYBP-PRC1 is recruited to target loci in mESCs and is also involved in Xist RNA-mediated silencing, the latter suggesting a wider role in Polycomb silencing. We discuss the implications of these findings for understanding recruitment and function of Polycomb repressors.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


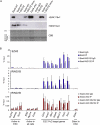


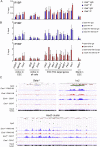
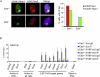
References
-
- Agger K., Cloos P.A.C., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. - PubMed
-
- Arrigoni R., Alam S.L., Wamstad J.A., Bardwell V.J., Sundquist W.I., Schreiber-Agus N. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006;580:6233–6241. - PubMed
-
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A.G. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Bejarano F., González I., Vidal M., Busturia A. The Drosophila RYBP gene functions as a Polycomb-dependent transcriptional repressor. Mech. Dev. 2005;122:1118–1129. - PubMed
-
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Supplemental References
-
- Atsuta, T., Fujimura, S., Moriya, H., Vidal, M., Akasaka, T., and Koseki, H. (2001). Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma 20, 43–46. - PubMed
-
- de Napoles, M., Mermoud, J.E., Wakao, R., Tang, Y.A., Endoh, M., Appanah, R., Nesterova, T.B., Silva, J., Otte, A.P., Vidal, M., et al. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676. - PubMed
-
- Dennis, G., Jr., Sherman, B.T., Hosack, D.A., Yang, J., Gao, W., Lane, H.C., and Lempicki, R.A. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, 3. - PubMed
-
- Huang, W., Sherman, B.T., and Lempicki, R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- G0900747/MRC_/Medical Research Council/United Kingdom
- WT085102MA/WT_/Wellcome Trust/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- 103768/WT_/Wellcome Trust/United Kingdom
- BBS/E/B/0000L748/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 087530/WT_/Wellcome Trust/United Kingdom
- 079249/WT_/Wellcome Trust/United Kingdom
- G0800784/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/0000H234/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0900747 91070/MRC_/Medical Research Council/United Kingdom
- 081385/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

