Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity
- PMID: 22325774
- PMCID: PMC3682648
- DOI: 10.1016/j.str.2011.11.017
Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity
Abstract
The cholesterol-dependent cytolysins (CDCs) punch holes in target cell membranes through a highly regulated process. Streptococcus mitis lectinolysin (LLY) exhibits another layer of regulation with a lectin domain that enhances the pore-forming activity of the toxin. We have determined the crystal structures of the lectin domain by itself and in complex with various glycans that reveal the molecular basis for the Lewis antigen specificity of LLY. A small-angle X-ray scattering study of intact LLY reveals the molecule is flat and elongated with the lectin domain oriented so that the Lewis antigen-binding site is exposed. We suggest that the lectin domain enhances the pore-forming activity of LLY by concentrating toxin molecules at fucose-rich sites on membranes, thus promoting the formation of prepore oligomers on the surface of susceptible cells.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
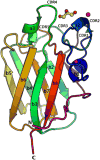
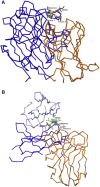
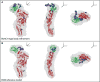
Comment in
-
Host glycan recognition by a pore forming toxin.Structure. 2012 Feb 8;20(2):197-8. doi: 10.1016/j.str.2012.01.013. Structure. 2012. PMID: 22325766 Free PMC article.
References
-
- Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. - PubMed
-
- Bianchet MA, Odom EW, Vasta GR, Amzel LM. A novel fucose recognition fold involved in innate immunity. Nat Struct Biol. 2002;9:628–634. - PubMed
-
- Boraston AB, Wang D, Burke RD. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006;281:35263–35271. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

