Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V
- PMID: 22326582
- PMCID: PMC3288297
- DOI: 10.1016/j.immuni.2011.12.014
Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V
Abstract
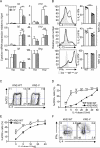
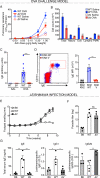
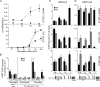
Follicular helper T cells (Tfh cells) are the major producers of interleukin-4 (IL-4) in secondary lymphoid organs where humoral immune responses develop. Il4 regulation in Tfh cells appears distinct from the classical T helper 2 (Th2) cell pathway, but the underlying molecular mechanisms remain largely unknown. We found that hypersensitivity site V (HS V; also known as CNS2), a 3' enhancer in the Il4 locus, is essential for IL-4 production by Tfh cells. Mice lacking HS V display marked defects in type 2 humoral immune responses, as evidenced by abrogated IgE and sharply reduced IgG1 production in vivo. In contrast, effector Th2 cells that are involved in tissue responses were far less dependent on HS V. HS V facilitated removal of repressive chromatin marks during Th2 and Tfh cell differentiation and increased accessibility of the Il4 promoter. Thus, Tfh and Th2 cells utilize distinct but overlapping molecular mechanisms to regulate Il4, a finding with important implications for understanding the molecular basis of allergic diseases.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. - PubMed
-
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. - PubMed
-
- Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. - PubMed
-
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

