Microbial functional gene diversity with a shift of subsurface redox conditions during In Situ uranium reduction
- PMID: 22327592
- PMCID: PMC3318829
- DOI: 10.1128/AEM.06528-11
Microbial functional gene diversity with a shift of subsurface redox conditions during In Situ uranium reduction
Abstract
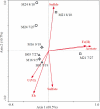
To better understand the microbial functional diversity changes with subsurface redox conditions during in situ uranium bioremediation, key functional genes were studied with GeoChip, a comprehensive functional gene microarray, in field experiments at a uranium mill tailings remedial action (UMTRA) site (Rifle, CO). The results indicated that functional microbial communities altered with a shift in the dominant metabolic process, as documented by hierarchical cluster and ordination analyses of all detected functional genes. The abundance of dsrAB genes (dissimilatory sulfite reductase genes) and methane generation-related mcr genes (methyl coenzyme M reductase coding genes) increased when redox conditions shifted from Fe-reducing to sulfate-reducing conditions. The cytochrome genes detected were primarily from Geobacter sp. and decreased with lower subsurface redox conditions. Statistical analysis of environmental parameters and functional genes indicated that acetate, U(VI), and redox potential (E(h)) were the most significant geochemical variables linked to microbial functional gene structures, and changes in microbial functional diversity were strongly related to the dominant terminal electron-accepting process following acetate addition. The study indicates that the microbial functional genes clearly reflect the in situ redox conditions and the dominant microbial processes, which in turn influence uranium bioreduction. Microbial functional genes thus could be very useful for tracking microbial community structure and dynamics during bioremediation.
Figures




References
-
- Boonchayaanant B, Nayak D, Xin D, Criddle C. 2009. Uranium reduction and resistance to reoxidation under iron-reducing and sulfate-reducing conditions. Water Res. 43:4652–4664 - PubMed
-
- Chang Y, et al. 2005. Microbial incorporation of C-13-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ. Sci. Technol. 39:9039–9048 - PubMed
-
- Clarke KR, Ainsworth M. 1993. A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92:205–219
-
- Druhan J, et al. 2008. Sulfur isotopes as indicators of amended bacterial sulfate reduction processes influencing field scale uranium bioremediation. Environ. Sci. Technol. 42:7842–7849 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

