EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock
- PMID: 22327739
- PMCID: PMC3315225
- DOI: 10.1105/tpc.111.093807
EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock
Abstract
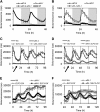
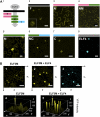
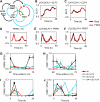
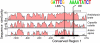
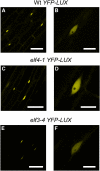
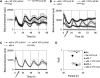
The plant circadian clock is proposed to be a network of several interconnected feedback loops, and loss of any component leads to changes in oscillator speed. We previously reported that Arabidopsis thaliana EARLY FLOWERING4 (ELF4) is required to sustain this oscillator and that the elf4 mutant is arrhythmic. This phenotype is shared with both elf3 and lux. Here, we show that overexpression of either ELF3 or LUX ARRHYTHMO (LUX) complements the elf4 mutant phenotype. Furthermore, ELF4 causes ELF3 to form foci in the nucleus. We used expression data to direct a mathematical position of ELF3 in the clock network. This revealed direct effects on the morning clock gene PRR9, and we determined association of ELF3 to a conserved region of the PRR9 promoter. A cis-element in this region was suggestive of ELF3 recruitment by the transcription factor LUX, consistent with both ELF3 and LUX acting genetically downstream of ELF4. Taken together, using integrated approaches, we identified ELF4/ELF3 together with LUX to be pivotal for sustenance of plant circadian rhythms.
Figures





References
-
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
-
- Brudno M., Malde S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S. (2003). Glocal alignment: finding rearrangements during alignment. Bioinformatics 19(suppl. 1): i54–i62 - PubMed
-
- Chen M. (2008). Phytochrome nuclear body: An emerging model to study interphase nuclear dynamics and signaling. Curr. Opin. Plant Biol. 11: 503–508 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

