Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins
- PMID: 22328055
- PMCID: PMC3332353
- DOI: 10.1007/s11103-012-9891-4
Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins
Abstract
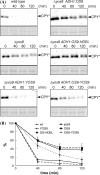
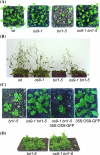
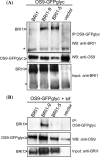
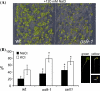
In the endoplasmic reticulum, immature polypeptides coincide with terminally misfolded proteins. Consequently, cells need a well-balanced quality control system, which decides about the fate of individual proteins and maintains protein homeostasis. Misfolded and unassembled proteins are sent for destruction via the endoplasmic reticulum-associated degradation (ERAD) machinery to prevent the accumulation of potentially toxic protein aggregates. Here, we report the identification of Arabidopsis thaliana OS9 as a component of the plant ERAD pathway. OS9 is an ER-resident glycoprotein containing a mannose-6-phosphate receptor homology domain, which is also found in yeast and mammalian lectins involved in ERAD. OS9 fused to the C-terminal domain of YOS9 can complement the ERAD defect of the corresponding yeast Δyos9 mutant. An A. thaliana OS9 loss-of-function line suppresses the severe growth phenotype of the bri1-5 and bri1-9 mutant plants, which harbour mutated forms of the brassinosteroid receptor BRI1. Co-immunoprecipitation studies demonstrated that OS9 associates with Arabidopsis SEL1L/HRD3, which is part of the plant ERAD complex and with the ERAD substrates BRI1-5 and BRI1-9, but only the binding to BRI1-5 occurs in a glycan-dependent way. OS9-deficiency results in activation of the unfolded protein response and reduces salt tolerance, highlighting the role of OS9 during ER stress. We propose that OS9 is a component of the plant ERAD machinery and may act specifically in the glycoprotein degradation pathway.
Figures




Similar articles
-
EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis.Proc Natl Acad Sci U S A. 2015 Sep 29;112(39):12205-10. doi: 10.1073/pnas.1511724112. Epub 2015 Sep 14. Proc Natl Acad Sci U S A. 2015. PMID: 26371323 Free PMC article.
-
The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases.Mol Plant. 2012 Jul;5(4):929-40. doi: 10.1093/mp/sss042. Epub 2012 Apr 19. Mol Plant. 2012. PMID: 22516478 Free PMC article.
-
Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis.Proc Natl Acad Sci U S A. 2011 Jan 11;108(2):870-5. doi: 10.1073/pnas.1013251108. Epub 2010 Dec 27. Proc Natl Acad Sci U S A. 2011. PMID: 21187394 Free PMC article.
-
The Role of Lectin-Carbohydrate Interactions in the Regulation of ER-Associated Protein Degradation.Molecules. 2015 May 27;20(6):9816-46. doi: 10.3390/molecules20069816. Molecules. 2015. PMID: 26023941 Free PMC article. Review.
-
The role of MRH domain-containing lectins in ERAD.Glycobiology. 2010 Jun;20(6):651-60. doi: 10.1093/glycob/cwq013. Epub 2010 Jan 28. Glycobiology. 2010. PMID: 20118070 Review.
Cited by
-
Endoplasmic reticulum-associated degradation of glycoproteins in plants.Front Plant Sci. 2012 Apr 5;3:67. doi: 10.3389/fpls.2012.00067. eCollection 2012. Front Plant Sci. 2012. PMID: 22645596 Free PMC article.
-
OS9 Protein Interacts with Na-K-2Cl Co-transporter (NKCC2) and Targets Its Immature Form for the Endoplasmic Reticulum-associated Degradation Pathway.J Biol Chem. 2016 Feb 26;291(9):4487-502. doi: 10.1074/jbc.M115.702514. Epub 2015 Dec 31. J Biol Chem. 2016. PMID: 26721884 Free PMC article.
-
EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis.Proc Natl Acad Sci U S A. 2015 Sep 29;112(39):12205-10. doi: 10.1073/pnas.1511724112. Epub 2015 Sep 14. Proc Natl Acad Sci U S A. 2015. PMID: 26371323 Free PMC article.
-
Biological significance of complex N-glycans in plants and their impact on plant physiology.Front Plant Sci. 2014 Jul 22;5:363. doi: 10.3389/fpls.2014.00363. eCollection 2014. Front Plant Sci. 2014. PMID: 25101107 Free PMC article. Review.
-
Protein Quality Control in the Endoplasmic Reticulum of Plants.Annu Rev Plant Biol. 2018 Apr 29;69:147-172. doi: 10.1146/annurev-arplant-042817-040331. Epub 2018 Mar 23. Annu Rev Plant Biol. 2018. PMID: 29570364 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

