Modification site localization scoring: strategies and performance
- PMID: 22328712
- PMCID: PMC3418845
- DOI: 10.1074/mcp.R111.015305
Modification site localization scoring: strategies and performance
Abstract
Using enrichment strategies many research groups are routinely producing large data sets of post-translationally modified peptides for proteomic analysis using tandem mass spectrometry. Although search engines are relatively effective at identifying these peptides with a defined measure of reliability, their localization of site/s of modification is often arbitrary and unreliable. The field continues to be in need of a widely accepted metric for false localization rate that accurately describes the certainty of site localization in published data sets and allows for consistent measurement of differences in performance of emerging scoring algorithms. In this article are discussed the main strategies currently used by software for modification site localization and ways of assessing the performance of these different tools. Methods for representing ambiguity are reviewed and a discussion of how the approaches transfer to different data types and modifications is presented.
Figures



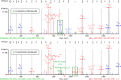
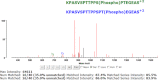
References
-
- Pawson T., Scott J. D. (2005) Protein phosphorylation in signaling–50 years and counting. Trends Biochem. Sci. 30, 286–290 - PubMed
-
- Boersema P. J., Mohammed S., Heck A. J. (2009) Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass Spectrom. 44, 861–878 - PubMed
-
- Hart G. W., Housley M. P., Slawson C. (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 - PubMed
-
- Rathert P., Dhayalan A., Ma H., Jeltsch A. (2008) Specificity of protein lysine methyltransferases and methods for detection of lysine methylation of non-histone proteins. Mol. Biosyst. 4, 1186–1190 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

