Spliceman--a computational web server that predicts sequence variations in pre-mRNA splicing
- PMID: 22328782
- PMCID: PMC3315715
- DOI: 10.1093/bioinformatics/bts074
Spliceman--a computational web server that predicts sequence variations in pre-mRNA splicing
Abstract
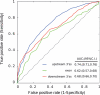
Summary: It was previously demonstrated that splicing elements are positional dependent. We exploited this relationship between location and function by comparing positional distributions between all possible 4096 hexamers around a database of human splice sites. The distance measure used in this study found point mutations that produced higher distances disrupted splicing, whereas point mutations with smaller distances generally had no effect on splicing. Reasoning the idea that functional splicing elements have signature positional distributions around constitutively spliced exons, we introduce Spliceman-an online tool that predicts how likely distant mutations around annotated splice sites were to disrupt splicing. Spliceman takes a set of DNA sequences with point mutations and returns a ranked list to predict the effects of point mutations on pre-mRNA splicing. The current implementation included the analyses of 11 genomes: human, chimp, rhesus, mouse, rat, dog, cat, chicken, guinea pig, frog and zebrafish.
Availability: Freely available on the web at http://fairbrother.biomed.brown.edu/spliceman/
Contact: fairbrother@brown.edu.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

