Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway
- PMID: 22330134
- PMCID: PMC3295664
- DOI: 10.1186/1475-2859-11-22
Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway
Abstract
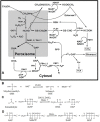
ΒACKGROUND: The methylotrophic yeast Pichia pastoris has become an important host organism for recombinant protein production and is able to use methanol as a sole carbon source. The methanol utilization pathway describes all the catalytic reactions, which happen during methanol metabolism. Despite the importance of certain key enzymes in this pathway, so far very little is known about possible effects of overexpressing either of these key enzymes on the overall energetic behavior, the productivity and the substrate uptake rate in P. pastoris strains.
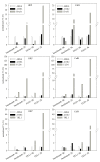
Results: A fast and easy-to-do approach based on batch cultivations with methanol pulses was used to characterize different P. pastoris strains. A strain with MutS phenotype was found to be superior over a strain with Mut+ phenotype in both the volumetric productivity and the efficiency in expressing recombinant horseradish peroxidase C1A. Consequently, either of the enzymes dihydroxyacetone synthase, transketolase or formaldehyde dehydrogenase, which play key roles in the methanol utilization pathway, was co-overexpressed in MutS strains harboring either of the reporter enzymes horseradish peroxidase or Candida antarctica lipase B. Although the co-overexpression of these enzymes did not change the stoichiometric yields of the recombinant MutS strains, significant changes in the specific growth rate, the specific substrate uptake rate and the specific productivity were observed. Co-overexpression of dihydroxyacetone synthase yielded a 2- to 3-fold more efficient conversion of the substrate methanol into product, but also resulted in a reduced volumetric productivity. Co-overexpression of formaldehyde dehydrogenase resulted in a 2-fold more efficient conversion of the substrate into product and at least similar volumetric productivities compared to strains without an engineered methanol utilization pathway, and thus turned out to be a valuable strategy to improve recombinant protein production.
Conclusions: Co-overexpressing enzymes of the methanol utilization pathway significantly affected the specific growth rate, the methanol uptake and the specific productivity of recombinant P. pastoris MutS strains. A recently developed methodology to determine strain specific parameters based on dynamic batch cultivations proved to be a valuable tool for fast strain characterization and thus early process development.
Figures
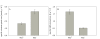

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

