Transcription as a source of genome instability
- PMID: 22330764
- PMCID: PMC3376450
- DOI: 10.1038/nrg3152
Transcription as a source of genome instability
Abstract
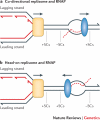
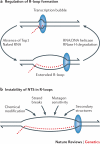
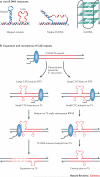
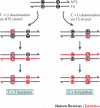
Alterations in genome sequence and structure contribute to somatic disease, affect the fitness of subsequent generations and drive evolutionary processes. The crucial roles of highly accurate replication and efficient repair in maintaining overall genome integrity are well-known, but the more localized stability costs that are associated with transcribing DNA into RNA molecules are less appreciated. Here we review the diverse ways in which the essential process of transcription alters the underlying DNA template and thereby modifies the genetic landscape.
Figures
References
-
- Savic DJ, Kanazir DT. The effect of a histidine operator-constitutive mutation on UV-induced mutability within the histidine operon of Salmonella typhimurium. Mol. Gen. Genet. 1972;118:45–50. - PubMed
-
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. [This work, using budding yeast as a model system, was the first demonstration of transcription-associated mutagenesis in eukaryotic cells.] - PubMed
-
- Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem. Rev. 2006;106:474–488. - PubMed
-
- Keil RL, Roeder GS. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

