Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein
- PMID: 22331412
- PMCID: PMC3320188
- DOI: 10.1104/pp.112.194035
Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein
Abstract
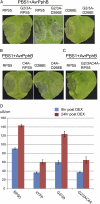
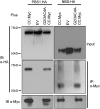
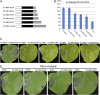
The Arabidopsis (Arabidopsis thaliana) RESISTANCE TO PSEUDOMONAS SYRINGAE5 (RPS5) disease resistance protein mediates recognition of the Pseudomonas syringae effector protein AvrPphB. RPS5 belongs to the coiled-coil-nucleotide-binding site-leucine-rich repeat (CC-NBS-LRR) family and is activated by AvrPphB-mediated cleavage of the protein kinase PBS1. Here, we present a structure-function analysis of the CC and LRR domains of RPS5 using transient expression assays in Nicotiana benthamiana. We found that substituting the CC domain of RPS2 for the RPS5 CC domain did not alter RPS5 specificity and only moderately reduced its ability to activate programmed cell death, suggesting that the CC domain does not play a direct role in the recognition of PBS1 cleavage. Analysis of an RPS5-super Yellow Fluorescent Protein fusion revealed that RPS5 localizes to the plasma membrane (PM). Alanine substitutions of predicted myristoylation (glycine-2) and palmitoylation (cysteine-4) residues affected RPS5 PM localization, protein stability, and function in an additive manner, indicating that PM localization is essential to RPS5 function. The first 20 amino acids of RPS5 were sufficient for directing super Yellow Fluorescent Protein to the PM. C-terminal truncations of RPS5 revealed that the first four LRR repeats are sufficient for inhibiting RPS5 autoactivation; however, the complete LRR domain was required for the recognition of PBS1 cleavage. Substitution of the RPS2 LRR domain resulted in the autoactivation of RPS5, indicating that the LRR domain must coevolve with the NBS domain. We conclude that the RPS5 LRR domain functions to suppress RPS5 activation in the absence of PBS1 cleavage and promotes RPS5 activation in its presence.
Figures






Similar articles
-
Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease.Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2531-6. doi: 10.1073/pnas.0608779104. Epub 2007 Feb 2. Proc Natl Acad Sci U S A. 2007. PMID: 17277084 Free PMC article.
-
Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on s-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1.Plant Physiol. 2014 Jan;164(1):340-51. doi: 10.1104/pp.113.227686. Epub 2013 Nov 13. Plant Physiol. 2014. PMID: 24225654 Free PMC article.
-
Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein.Cell Microbiol. 2012 Jul;14(7):1071-84. doi: 10.1111/j.1462-5822.2012.01779.x. Epub 2012 Mar 27. Cell Microbiol. 2012. PMID: 22372664 Free PMC article.
-
RPS5-Mediated Disease Resistance: Fundamental Insights and Translational Applications.Annu Rev Phytopathol. 2020 Aug 25;58:139-160. doi: 10.1146/annurev-phyto-010820-012733. Epub 2020 Apr 13. Annu Rev Phytopathol. 2020. PMID: 32284014 Review.
-
Effector recognition and activation of the Arabidopsis thaliana NLR innate immune receptors.Cold Spring Harb Symp Quant Biol. 2012;77:249-57. doi: 10.1101/sqb.2012.77.014860. Epub 2012 Nov 21. Cold Spring Harb Symp Quant Biol. 2012. PMID: 23174767 Review.
Cited by
-
Recent Advances in Effector-Triggered Immunity in Plants: New Pieces in the Puzzle Create a Different Paradigm.Int J Mol Sci. 2021 Apr 29;22(9):4709. doi: 10.3390/ijms22094709. Int J Mol Sci. 2021. PMID: 33946790 Free PMC article. Review.
-
Plant NLR immune receptor Tm-22 activation requires NB-ARC domain-mediated self-association of CC domain.PLoS Pathog. 2020 Apr 27;16(4):e1008475. doi: 10.1371/journal.ppat.1008475. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32339200 Free PMC article.
-
A maize polygalacturonase functions as a suppressor of programmed cell death in plants.BMC Plant Biol. 2019 Jul 15;19(1):310. doi: 10.1186/s12870-019-1897-5. BMC Plant Biol. 2019. PMID: 31307401 Free PMC article.
-
The role of lipid post-translational modification in plant developmental processes.Front Plant Sci. 2014 Feb 18;5:50. doi: 10.3389/fpls.2014.00050. eCollection 2014. Front Plant Sci. 2014. PMID: 24600462 Free PMC article. Review.
-
Evolutionary relationship of disease resistance genes in soybean and Arabidopsis specific for the Pseudomonas syringae effectors AvrB and AvrRpm1.Plant Physiol. 2014 Sep;166(1):235-51. doi: 10.1104/pp.114.244715. Epub 2014 Jul 17. Plant Physiol. 2014. PMID: 25034017 Free PMC article.
References
-
- Albrecht M, Takken FL. (2006) Update on the domain architectures of NLRs and R proteins. Biochem Biophys Res Commun 339: 459–462 - PubMed
-
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 - PubMed
-
- Axtell MJ, Staskawicz BJ. (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

