A role for sphingomyelin-rich lipid domains in the accumulation of phosphatidylinositol-4,5-bisphosphate to the cleavage furrow during cytokinesis
- PMID: 22331463
- PMCID: PMC3318597
- DOI: 10.1128/MCB.06113-11
A role for sphingomyelin-rich lipid domains in the accumulation of phosphatidylinositol-4,5-bisphosphate to the cleavage furrow during cytokinesis
Abstract
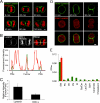
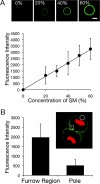
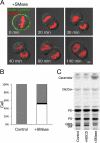
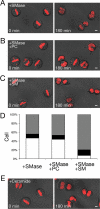
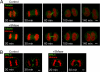
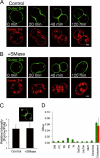
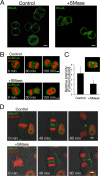
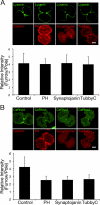
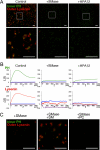
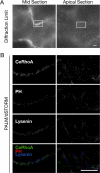
Cytokinesis is a crucial step in the creation of two daughter cells by the formation and ingression of the cleavage furrow. Here, we show that sphingomyelin (SM), one of the major sphingolipids in mammalian cells, is required for the localization of phosphatidylinositol-4,5-bisphosphate (PIP(2)) to the cleavage furrow during cytokinesis. Real-time observation with a labeled SM-specific protein, lysenin, revealed that SM is concentrated in the outer leaflet of the furrow at the time of cytokinesis. Superresolution fluorescence microscopy analysis indicates a transbilayer colocalization between the SM-rich domains in the outer leaflet and PIP(2)-rich domains in the inner leaflet of the plasma membrane. The depletion of SM disperses PIP(2) and inhibits the recruitment of the small GTPase RhoA to the cleavage furrow, leading to abnormal cytokinesis. These results suggest that the formation of SM-rich domains is required for the accumulation of PIP(2) to the cleavage furrow, which is a prerequisite for the proper translocation of RhoA and the progression of cytokinesis.
Figures



References
-
- Ando R, Mizuno H, Miyawaki A. 2004. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306: 1370–1373 - PubMed
-
- Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 - PubMed
-
- Brown DA, London E. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14: 111–136 - PubMed
-
- Echard A. 2008. Membrane traffic and polarization of lipid domains during cytokinesis. Biochem. Soc. Trans. 36: 395–399 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
