Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore
- PMID: 22331849
- PMCID: PMC3284002
- DOI: 10.1083/jcb.201111107
Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore
Abstract
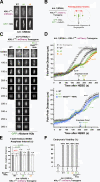
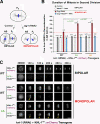
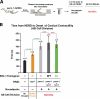
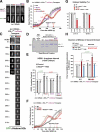
Accurate chromosome segregation requires coordination between microtubule attachment and spindle checkpoint signaling at the kinetochore. The kinetochore-localized KMN (KNL-1/Mis12 complex/Ndc80 complex) network, which mediates microtubule attachment and scaffolds checkpoint signaling, harbors two distinct microtubule-binding activities: the load-bearing activity of the Ndc80 complex and a less well-understood activity in KNL-1. In this paper, we show that KNL-1 microtubule-binding and -bundling activity resides in its extreme N terminus. Selective perturbation of KNL-1 microtubule binding in Caenorhabditis elegans embryos revealed that this activity is dispensable for both load-bearing attachment formation and checkpoint activation but plays a role in checkpoint silencing at the kinetochore. Perturbation of both microtubule binding and protein phosphatase 1 docking at the KNL-1 N terminus additively affected checkpoint silencing, indicating that, despite their proximity in KNL-1, these two activities make independent contributions. We propose that microtubule binding by KNL-1 functions in checkpoint silencing by sensing microtubules attached to kinetochores and relaying their presence to eliminate generation of the checkpoint signal.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

