The p53 circuit board
- PMID: 22333261
- PMCID: PMC3307870
- DOI: 10.1016/j.bbcan.2012.01.004
The p53 circuit board
Abstract
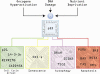
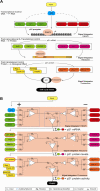
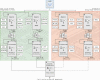
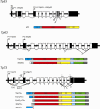
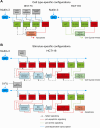
The p53 tumor suppressor is embedded in a large gene network controlling diverse cellular and organismal phenotypes. Multiple signaling pathways converge onto p53 activation, mostly by relieving the inhibitory effects of its repressors, MDM2 and MDM4. In turn, signals originating from increased p53 activity diverge into distinct effector pathways to deliver a specific cellular response to the activating stimuli. Much attention has been devoted to dissecting how the various input pathways trigger p53 activation and how the activity of the p53 protein itself can be modulated by a plethora of co-factors and post-translational modifications. In this review we will focus instead on the multiple configurations of the effector pathways. We will discuss how p53-generated signals are transmitted, amplified, resisted and eventually integrated by downstream gene circuits operating at the transcriptional, post-transcriptional and post-translational levels. We will also discuss how context-dependent variations in these gene circuits define the cellular response to p53 activation and how they may impact the clinical efficacy of p53-based targeted therapies.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures

References
-
- Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. - PubMed
-
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. - PubMed
-
- Eliyahu D, Raz A, Gruss P, Givol D, Oren M. Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature. 1984;312:646–649. - PubMed
-
- Jenkins JR, Rudge K, Currie GA. Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature. 1984;312:651–654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

