Yabi: An online research environment for grid, high performance and cloud computing
- PMID: 22333270
- PMCID: PMC3298538
- DOI: 10.1186/1751-0473-7-1
Yabi: An online research environment for grid, high performance and cloud computing
Abstract
Background: There is a significant demand for creating pipelines or workflows in the life science discipline that chain a number of discrete compute and data intensive analysis tasks into sophisticated analysis procedures. This need has led to the development of general as well as domain-specific workflow environments that are either complex desktop applications or Internet-based applications. Complexities can arise when configuring these applications in heterogeneous compute and storage environments if the execution and data access models are not designed appropriately. These complexities manifest themselves through limited access to available HPC resources, significant overhead required to configure tools and inability for users to simply manage files across heterogenous HPC storage infrastructure.
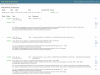
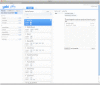
Results: In this paper, we describe the architecture of a software system that is adaptable to a range of both pluggable execution and data backends in an open source implementation called Yabi. Enabling seamless and transparent access to heterogenous HPC environments at its core, Yabi then provides an analysis workflow environment that can create and reuse workflows as well as manage large amounts of both raw and processed data in a secure and flexible way across geographically distributed compute resources. Yabi can be used via a web-based environment to drag-and-drop tools to create sophisticated workflows. Yabi can also be accessed through the Yabi command line which is designed for users that are more comfortable with writing scripts or for enabling external workflow environments to leverage the features in Yabi. Configuring tools can be a significant overhead in workflow environments. Yabi greatly simplifies this task by enabling system administrators to configure as well as manage running tools via a web-based environment and without the need to write or edit software programs or scripts. In this paper, we highlight Yabi's capabilities through a range of bioinformatics use cases that arise from large-scale biomedical data analysis.
Conclusion: The Yabi system encapsulates considered design of both execution and data models, while abstracting technical details away from users who are not skilled in HPC and providing an intuitive drag-and-drop scalable web-based workflow environment where the same tools can also be accessed via a command line. Yabi is currently in use and deployed at multiple institutions and is available at http://ccg.murdoch.edu.au/yabi.
Figures
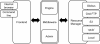


References
-
- Louys M, Bonnarel F, Schaaff A, Claudon J-J, Pestel C. In: Highlights of Astronomy, XXVIth IAU General Assembly. van der Hucht KA, editor. Vol. 14 2006. Implementing astronomical image analysis pipelines using VO standards.
-
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehväslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12(10):1611–8. doi: 10.1101/gr.361602. - DOI - PMC - PubMed
-
- Pocock M, Down T, Hubbard T. BioJava: open source components for bioinformatics. ACM SIGBIO Newsletter. 2000;20(2):10–12. doi: 10.1145/360262.360266. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

