A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis
- PMID: 22333914
- PMCID: PMC3321182
- DOI: 10.1038/emboj.2012.26
A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis
Abstract
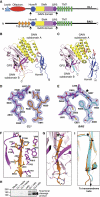
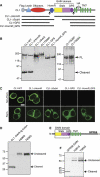
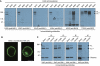
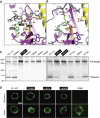
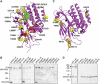
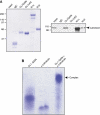
The G protein-coupled receptor (GPCR) Proteolysis Site (GPS) of cell-adhesion GPCRs and polycystic kidney disease (PKD) proteins constitutes a highly conserved autoproteolysis sequence, but its catalytic mechanism remains unknown. Here, we show that unexpectedly the ∼40-residue GPS motif represents an integral part of a much larger ∼320-residue domain that we termed GPCR-Autoproteolysis INducing (GAIN) domain. Crystal structures of GAIN domains from two distantly related cell-adhesion GPCRs revealed a conserved novel fold in which the GPS motif forms five β-strands that are tightly integrated into the overall GAIN domain. The GAIN domain is evolutionarily conserved from tetrahymena to mammals, is the only extracellular domain shared by all human cell-adhesion GPCRs and PKD proteins, and is the locus of multiple human disease mutations. Functionally, the GAIN domain is both necessary and sufficient for autoproteolysis, suggesting an autoproteolytic mechanism whereby the overall GAIN domain fine-tunes the chemical environment in the GPS to catalyse peptide bond hydrolysis. Thus, the GAIN domain embodies a unique, evolutionarily ancient and widespread autoproteolytic fold whose function is likely relevant for GPCR signalling and for multiple human diseases.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

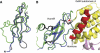
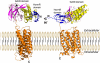
Comment in
-
A GAIN in understanding autoproteolytic G protein-coupled receptors and polycystic kidney disease proteins.EMBO J. 2012 Mar 21;31(6):1334-5. doi: 10.1038/emboj.2012.51. Epub 2012 Mar 2. EMBO J. 2012. PMID: 22388517 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 - PMC - PubMed
-
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, Domene S, Velez JI, Karkera JD, Balog J, Berg K, Kleta R, Gahl WA, Roessler E, Long R, Lie J, Pineda D, Londono AC, Palacio JD, Arbelaez A et al. (2010) A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry 15: 1053–1066 - PubMed
-
- Blobel CP (2005) ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol 6: 32–43 - PubMed
-
- Chae J, Kim MJ, Goo JH, Collier S, Gubb D, Charlton J, Adler PN, Park WJ (1999) The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development 126: 5421–5429 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

