Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life
- PMID: 22334342
- PMCID: PMC3376375
- DOI: 10.1093/sysbio/sys026
Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life
Abstract
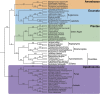
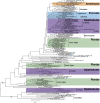
The first analyses of gene sequence data indicated that the eukaryotic tree of life consisted of a long stem of microbial groups "topped" by a crown-containing plants, animals, and fungi and their microbial relatives. Although more recent multigene concatenated analyses have refined the relationships among the many branches of eukaryotes, the root of the eukaryotic tree of life has remained elusive. Inferring the root of extant eukaryotes is challenging because of the age of the group (∼1.7-2.1 billion years old), tremendous heterogeneity in rates of evolution among lineages, and lack of obvious outgroups for many genes. Here, we reconstruct a rooted phylogeny of extant eukaryotes based on minimizing the number of duplications and losses among a collection of gene trees. This approach does not require outgroup sequences or assumptions of orthology among sequences. We also explore the impact of taxon and gene sampling and assess support for alternative hypotheses for the root. Using 20 gene trees from 84 diverse eukaryotic lineages, this approach recovers robust eukaryotic clades and reveals evidence for a eukaryotic root that lies between the Opisthokonta (animals, fungi and their microbial relatives) and all remaining eukaryotes.
Figures
References
-
- Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup Ø, Mozley-Standrdge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MFJR. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 2005;52:399–451. - PubMed
-
- Arisue N, Hasegawa M, Hashimoto T. Root of the eukaryota tree as inferred from combined maximum likelihood analyses of multiple molecular sequence data. Mol. Biol. Evol. 2005;22:409–420. - PubMed
-
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. - PubMed
-
- Brinkmann H, Philippe H. Berlin (Germany): Springer-Verlag; 2007. The diversity of eukaryotes and the root of the eukaryotic tree. Eukaryotic membranes and cytoskeleton: origins and evolution; pp. p. 20–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

