Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy
- PMID: 22334415
- PMCID: PMC3314127
- DOI: 10.1002/ana.22683
Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy
Abstract
Objective: To identify the causative gene in an autosomal dominant limb-girdle muscular dystrophy (LGMD) with skeletal muscle vacuoles.
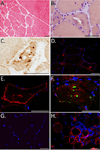
Methods: Exome sequencing was used to identify candidate mutations in the studied pedigree. Genome-wide linkage was then used to narrow the list of candidates to a single disease-associated mutation. Additional pedigrees with dominant or sporadic myopathy were screened for mutations in the same gene (DNAJB6) using exome sequencing. Skeletal muscle from affected patients was evaluated with histochemistry and immunohistochemical stains for dystrophy-related proteins, SMI-31, TDP43, and DNAJB6.
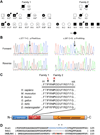
Results: Exome analysis in 3 affected individuals from a family with dominant LGMD and vacuolar pathology identified novel candidate mutations in 22 genes. Linkage analysis excluded all variants except a Phe93Leu mutation in the G/F domain of the DNAJB6 gene, which resides within the LGMD locus at 7q36. Analysis of exome sequencing data from other pedigrees with dominant myopathy identified a second G/F domain mutation (Pro96Arg) in DNAJB6. Affected muscle showed mild dystrophic changes, vacuoles, and abnormal aggregation of proteins, including TDP-43 and DNAJB6 itself.
Interpretation: Mutations within the G/F domain of DNAJB6 are a novel cause of dominantly-inherited myopathy. DNAJB6 is a member of the HSP40/DNAJ family of molecular co-chaperones tasked with protecting client proteins from irreversible aggregation during protein synthesis or during times of cellular stress. The abnormal accumulation of several proteins in patient muscle, including DNAJB6 itself, suggest that DNAJB6 function is compromised by the identified G/F domain mutations.
Copyright © 2012 American Neurological Association.
Figures
References
-
- Bushby KM. Diagnostic criteria for the limb-girdle muscular dystrophies: report of the ENMC Consortium on Limb-Girdle Dystrophies. Neuromuscul Disord. 1995 Jan;5(1):71–74. - PubMed
-
- Pestronk A. [cited 2011 July 22];Limb-girdle muscular dystrophy (LGMD) Syndromes. 2011 Available from: http://neuromuscular.wustl.edu.
-
- Hauser MA, Horrigan SK, Salmikangas P, et al. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum Mol Genet. 2000 Sep 1;9(14):2141–2147. - PubMed
-
- Muchir A, Bonne G, van der Kooi AJ, et al. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum Mol Genet. 2000 May 22;9(9):1453–1459. - PubMed
-
- Minetti C, Sotgia F, Bruno C, et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998 Apr;18(4):365–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 NS057105/NS/NINDS NIH HHS/United States
- K08 NS055980/NS/NINDS NIH HHS/United States
- K08 NS075094/NS/NINDS NIH HHS/United States
- NS057105/NS/NINDS NIH HHS/United States
- NS075094/NS/NINDS NIH HHS/United States
- R01 NS069669/NS/NINDS NIH HHS/United States
- AG031867/AG/NIA NIH HHS/United States
- NS069669/NS/NINDS NIH HHS/United States
- NS055980/NS/NINDS NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- R01 AG031867/AG/NIA NIH HHS/United States
- UL1RR024992/RR/NCRR NIH HHS/United States
- UL1 RR024992/RR/NCRR NIH HHS/United States
- P30 CA91842/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

