Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia
- PMID: 22334516
- PMCID: PMC3280448
- DOI: 10.1128/mBio.00240-11
Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia
Abstract
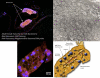
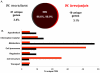
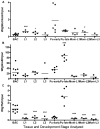
Ancient endosymbionts have been associated with extreme genome structural stability with little differentiation in gene inventory between sister species. Tsetse flies (Diptera: Glossinidae) harbor an obligate endosymbiont, Wigglesworthia, which has coevolved with the Glossina radiation. We report on the ~720-kb Wigglesworthia genome and its associated plasmid from Glossina morsitans morsitans and compare them to those of the symbiont from Glossina brevipalpis. While there was overall high synteny between the two genomes, a large inversion was noted. Furthermore, symbiont transcriptional analyses demonstrated host tissue and development-specific gene expression supporting robust transcriptional regulation in Wigglesworthia, an unprecedented observation in other obligate mutualist endosymbionts. Expression and immunohistochemistry confirmed the role of flagella during the vertical transmission process from mother to intrauterine progeny. The expression of nutrient provisioning genes (thiC and hemH) suggests that Wigglesworthia may function in dietary supplementation tailored toward host development. Furthermore, despite extensive conservation, unique genes were identified within both symbiont genomes that may result in distinct metabolomes impacting host physiology. One of these differences involves the chorismate, phenylalanine, and folate biosynthetic pathways, which are uniquely present in Wigglesworthia morsitans. Interestingly, African trypanosomes are auxotrophs for phenylalanine and folate and salvage both exogenously. It is possible that W. morsitans contributes to the higher parasite susceptibility of its host species.
Importance: Genomic stasis has historically been associated with obligate endosymbionts and their sister species. Here we characterize the Wigglesworthia genome of the tsetse fly species Glossina morsitans and compare it to its sister genome within G. brevipalpis. The similarity and variation between the genomes enabled specific hypotheses regarding functional biology. Expression analyses indicate significant levels of transcriptional regulation and support development- and tissue-specific functional roles for the symbiosis previously not observed in obligate mutualist symbionts. Retention of the genetically expensive flagella within these small genomes was demonstrated to be significant in symbiont transmission and tailored to the unique tsetse fly reproductive biology. Distinctions in metabolomes were also observed. We speculate an additional role for Wigglesworthia symbiosis where infections with pathogenic trypanosomes may depend upon symbiont species-specific metabolic products and thus influence the vector competence traits of different tsetse fly host species.
Figures



References
-
- Buchner P. 1965. Endosymbiosis of animals with plant microorganisms, p. 210–338 Interscience Publishing Group Inc, New York, NY.
-
- Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 - PubMed
-
- Clark M, Moran N, Baumann P. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol. Biol. Evol. 16:1586–1598 - PubMed
-
- Wernegreen JJ. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850–861 - PubMed
-
- Moran NA, McLaughlin HJ, Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

