Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons
- PMID: 22334647
- PMCID: PMC3295291
- DOI: 10.1073/pnas.1108718109
Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons
Abstract
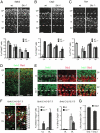
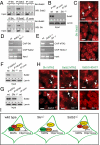
First insights into the molecular programs orchestrating the progression from neural stem cells to cortical projection neurons are emerging. Loss of the transcriptional regulator Ski has been linked to the human 1p36 deletion syndrome, which includes central nervous system defects. Here, we report critical roles for Ski in the maintenance of the neural stem cell pool and the specification of callosal neurons. Ski-deficient callosal neurons lose their identity and ectopically express the transcription factor Ctip2. The misspecified callosal neurons largely fail to form the corpus callosum and instead redirect their axons toward subcortical targets. We identify the chromatin-remodeling factor Satb2 as a partner of Ski, and show that both proteins are required for transcriptional repression of Ctip2 in callosal neurons. We propose a model in which Satb2 recruits Ski to the Ctip2 locus, and Ski attracts histone deacetylases, thereby enabling the formation of a functional nucleosome remodeling and deacetylase repressor complex. Our findings establish a central role for Ski-Satb2 interactions in regulating transcriptional mechanisms of callosal neuron specification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. - PubMed
-
- Bonnon C, Atanasoski S. c-Ski in health and disease. Cell Tissue Res. 2012;347:51–64. - PubMed
-
- Lyons GE, et al. Protooncogene c-Ski is expressed in both proliferating and postmitotic neuronal populations. Dev Dyn. 1994;201:354–365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

